🧬 GEBRA™ Use Guide: Knowledge Base – Interpretations That Persist, Diagnoses That Compound

🧬 “I’ve seen this variant before… where was it?”
It happens all the time.
You notice a variant, you’re sure you’ve seen it in another patient, then you start searching—papers, spreadsheets, a local DB, ClinVar—minutes turn into half an hour. And even if you find it, you still have to check the phenotype context.
GEBRA Knowledge Base ends that chase with a single search.
🧠 Three Databases. One Platform.
Together, these three resources raise both speed and depth of your work:
| Database | Role | Best used when… |
| CurationDB | Your institution’s curated interpretation notes per variant (comments, evidence, citations). | You want to preserve and reuse your own reasoning. |
| VariantsDB | Every variant observed during analyses (not only reported ones). | You need a quick look-up of where a variant has appeared across samples. |
| ReportedDB | The subset of variants that were actually reported. | You need audit-ready history, QC, or follow-up planning. |
In short, GEBRA turns experience into structured, searchable data.
🔍 Find what you need—instantly
VariantsDB
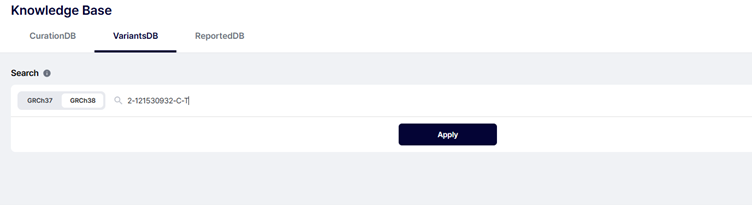
Enter a genomic position and see all samples where it appears.
Try: 2-121530932-C-T
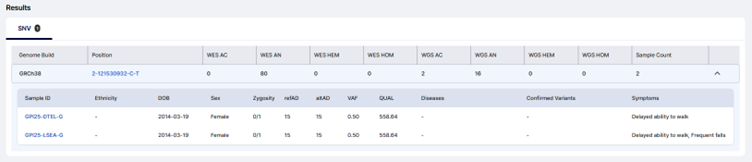
This isn’t just a hit list. You can jump to each sample’s phenotype, prior interpretation context, and report status—all centered on the same variant.
Same variant, different patients.
That comparison often unlocks new interpretation.
ReportedDB
Need your institution-level history?
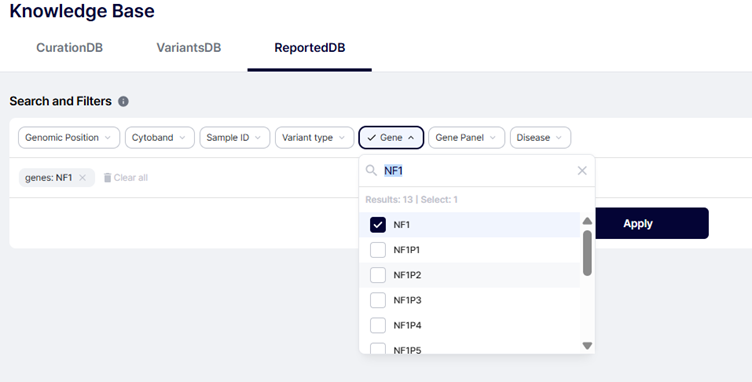
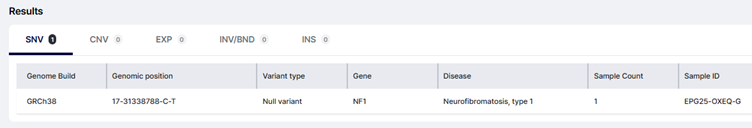
Search by gene—e.g., NF1—and you’ll see all reported NF1 variants to date, with genomic coordinates, HGVS, ACMG classification, and links back to the original cases.
CurationDB
Log your judgment where it matters: at the variant.
Attach paper links, rationale for ACMG criteria, segregation notes, internal comments, and institutional conventions.
This isn’t a personal notebook—it’s institutional memory.
GEBRA turns individual expertise into data that compounds over time.
⚙️ Knowledge connected, diagnosis accelerated
The real strength of Knowledge Base isn’t just per-patient lookup.
It’s spotting patterns across your cohort:
- Search SLC25A1 in VariantsDB and see it in five different patients with varied phenotypes.
- Identify recurrence, check zygosity, compare clinical contexts.
- Decide whether a VUS stays uncertain—or now has direction for reclassification.
By reusing prior cases, you shorten time-to-diagnosis and increase consistency and confidence.
🧭 Keep the context of your judgment
Outcomes matter—but why you decided matters more.
Knowledge Base preserves rationale, references, and meeting notes so you can always answer: “Why did we call it that way?”
This supports molecular board prep, QC.
Not just data accumulation—context preservation for defensible decisions.
🧪 How value compounds over time
- 1 month later: “We saw this variant last week with an unrelated phenotype—benign is plausible.”
- 6 months later: “Five occurrences in our population; likely benign here, even if public DBs still label it rare.”
- 12 months later: “This VUS appeared in three more patients with matching phenotypes and segregation—now reclassify to LP with confidence.”
Your data becomes your differentiator.
Knowledge Base ensures it’s never lost and always usable.
🚀 Getting started
No special setup:
- Running an analysis auto-populates VariantsDB.
- Issuing a report auto-updates ReportedDB.
- Add your notes to CurationDB whenever you interpret.
On your GEBRA, open Knowledge Base from the left menu and start searching—by gene, genomic position, disease, panel, or classification.
📍 Learn More
Explore how GEBRA integrates automation with clinical expertise.
Take a look at all GEBRA™ Related Articles:
- GEBRA™ Use Guide: 3ASC – Finding the One Causal Variant Among Thousands
- GEBRA™ Use Guide: Symptom-driven Update — When Phenotypes Evolve, Your Shortlist Should Too
- GEBRA™ Use Guide: Filters – See What Matters, Faster
- GEBRA™ Use Guide: Knowledge Base – Interpretations That Persist, Diagnoses That Compound
- GEBRA™ Use Guide: Gene Coverage – Diagnostic Confidence Starts with Coverage
- GEBRA™ Use Guide: 3bCNV – From Copy Number Variant Detection to Clinical Interpretation
- GEBRA™ Use Guide: Symptom Color Tag
Get exclusive rare disease updates
from 3billion.

3billion Inc.
3billion is dedicated to creating a world where patients with rare diseases are not neglected in diagnosis and treatment.






