Can Exome Sequencing Detect CNVs?

When a patient presents with symptoms suggestive of a rare genetic disorder, which diagnostic test comes to mind first?
For many clinicians, chromosomal microarray (CMA) is still the go-to, especially for detecting copy number variants (CNVs). CNVs—defined as deletions or duplications of 50 base pairs or more—are implicated in approximately 10–15% of genetic disorders. CMA has long been used as the gold standard to identify these larger genomic changes, capable of detecting CNVs greater than 50 kilobases (kb) in size.
However, CMA isn’t without its limitations. It cannot reliably detect:
- CNVs smaller than 50kb, and
- SNVs accompanied by CNVs in trans, which can lead to undiagnosed autosomal recessive conditions.
So, where does exome sequencing fit into this picture?
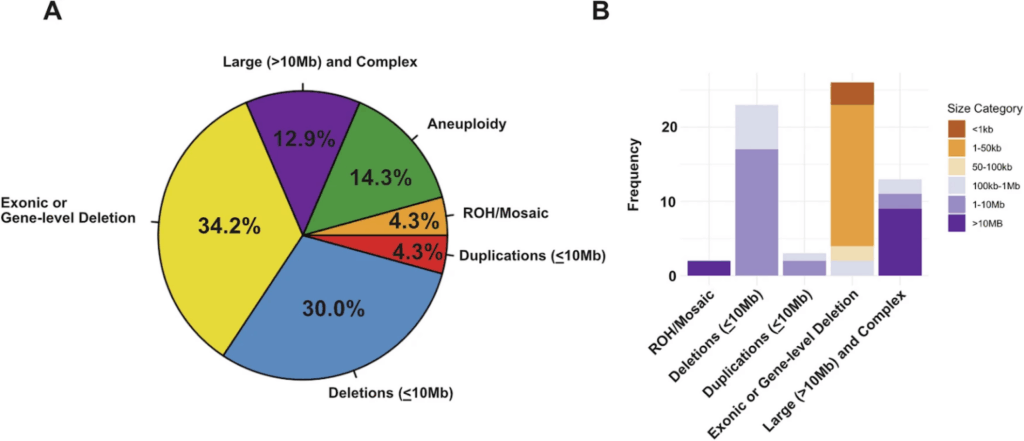
Visual data shows that the orange and dark orange zones in CNV size distribution are where exome sequencing outperforms CMA—areas where diagnostic opportunities would otherwise be lost.
Can it replace CMA?
The Short Answer: Yes—with one important caveat.
Exome sequencing can effectively detect CNVs, as long as the CNV spans three or more consecutive exons. This makes it a powerful diagnostic tool for identifying smaller, gene-level CNVs that CMA may miss.
In contrast, CMA can detect even a single-exon CNV, but only if that exon is larger than 50kb—something that’s rare in the human genome. So, in cases where a single small exon is involved, CMA might actually succeed where standard-depth exome sequencing does not.
But here’s the solution: increasing the read depth of exome sequencing from 100x to 300x significantly improves its sensitivity, even for single-exon deletions or duplications.
What the Research Says
A February 2025 study published in npj Genomic Medicine found that high-depth exome sequencing:
- Detected pathogenic CNVs under 50kb that CMA completely missed, and
- Identified trans SNVs in combination with CNVs, improving diagnostic accuracy in autosomal recessive disorders.
This dual advantage significantly enhances the overall diagnostic yield and could reduce the diagnostic odyssey many patients face—saving both time and cost.
What This Means for Clinical Practice
If you’re relying solely on CMA for CNV analysis, you might be missing important findings. Exome sequencing not only broadens your detection range but also integrates SNV and CNV data in one test.
Here’s a breakdown of what exome sequencing can detect:
- CNV Categories: Multi-exon deletions/duplications, especially 3+ exons
- Smaller CNVs missed by CMA: Particularly those under 50kb
- SNV + CNV combinations: Captured more comprehensively than by CMA
Do you have a case where CNV detection was inconclusive?
Reach out to us you here, and we’ll support you with a complementary trial.
Let’s improve diagnostic confidence—together.
Get exclusive rare disease updates
from 3billion.

Sree Ramya Gunukula
Marketing Leader with experience in the pharma and healthcare sectors, specializing in digital health, genetic testing, and rare disease diagnostics.







