Software that learn come in many forms and are already in our lives. If you’ve ever used Google Translate, spoken to Siri, driven a Tesla (using Autopilot) or made a credit card transaction, you have either directly or indirectly interacted with AI. Just as AI improved accessibility and convenience in day-to-day applications, its potential benefits are also promising in rare genetic disease diagnosis.
But there is a catch -- despite all the effort made to ensure that the AI is accurate, what happens when it’s wrong? The truth is often complex, and this case is no different: it depends. The applications mentioned above either cause little more than user disappointment when wrong (Google Translate and Siri), or delegate the final decision to humans (fraud detection; also, a reminder that Autopilot1 is SAE level 22). Along the same vein, the most immediately adoptable applications of AI in rare disease diagnosis are ones that are the safest and relatively lower in impact.

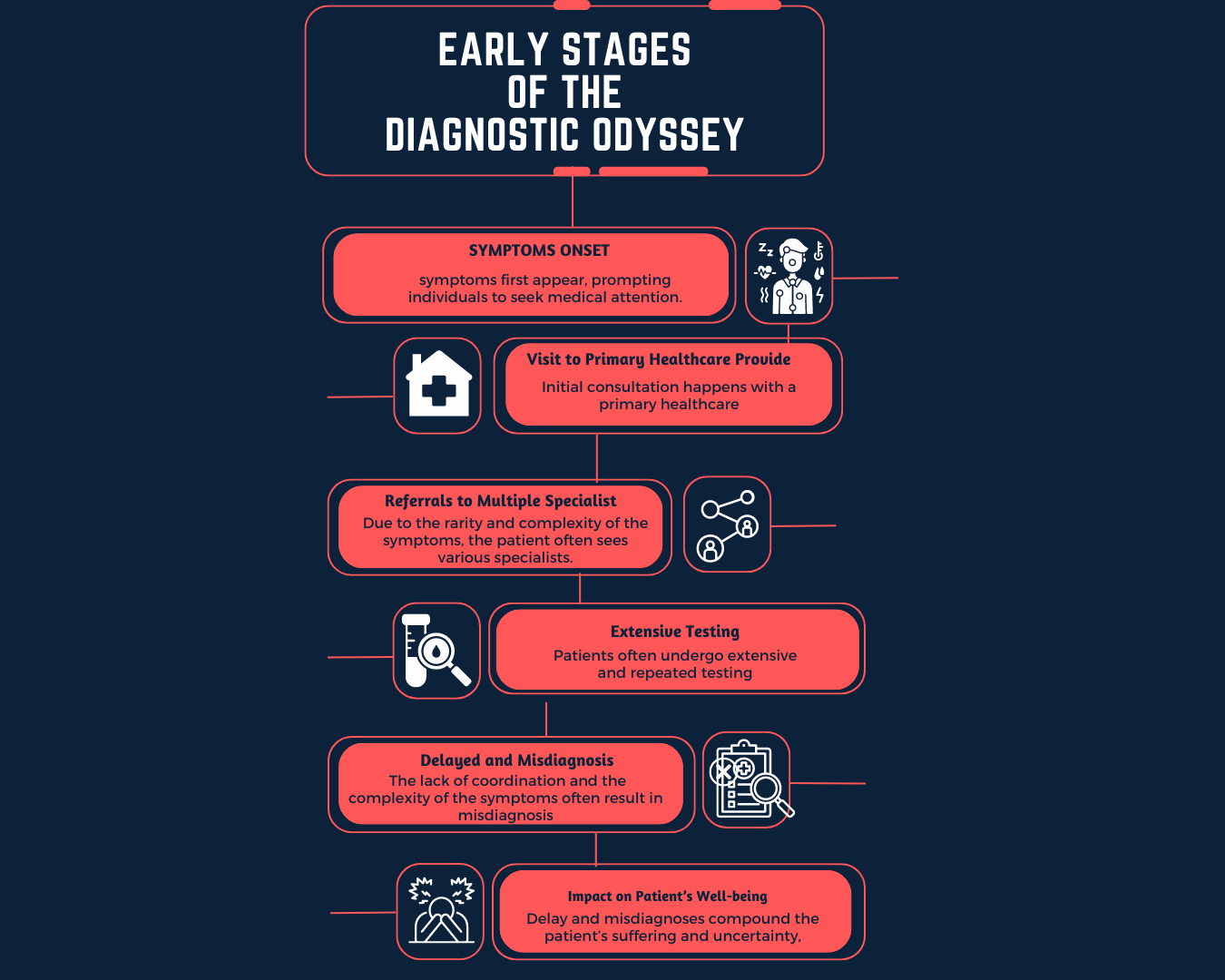
How could AI help in the diagnosis of rare diseases?
In a field where an accepted solution already exists, AI is introduced with the intention to cut costs, further improve results, or in more ideal cases, do both. Listed below are some possible applications of AI in the rare disease diagnosis space.
Phenotype suggestions and detection from patient descriptions
Standardization is particularly important in diagnostic pipelines with automated components, and a feature that reads over patient descriptions then suggests phenotype keywords prior to genetic test order submission would be a helpful but minimally-disruptive application of AI in rare disease diagnosis. All notable and relevant patient phenotypes should be submitted when requesting a genetic test. (Not familiar with phenotyping and its role in diagnosis? This piece is a good place to start.) While it is preferred that phenotypes are documented in terms of the Human Phenotype Ontology (HPO)3, an organized vocabulary used to describe phenotypes and diseases, patient phenotypes are sometimes written as free-form descriptions, which sometimes introduce semantic ambiguities and take longer to process.
The automated detection of patient phenotypes is expected to shorten case processing times by about five minutes per case and reduce the amount of mental exertion on clinical geneticists. A description of the patient’s phenotype is essential for proper genetic testing, and given that everything else is equal, cases with more relevant and precise descriptions have a higher probability of diagnosis relative to cases with fewer or imprecise ones. By suggesting other associated phenotypes based on ones already provided, healthcare professionals have the opportunity to consider unexplored possibilities before submitting their request for genetic testing. Considering that the diagnosis rate for cases with one phenotype provided is 40% and those with ten or more is 57.7%4, the discovery of additional patient-specific phenotypes leads to higher rates of diagnosis and opportunities for treatment.
Automated curation and tagging of publications regarding genetic diseases
Diagnoses are made based on published research that connect mutations to symptoms, and ultimately to diseases. The field of genetic disease is ever evolving; with thousands of works of research becoming newly available daily, but with only a small portion of them pertaining to genetic diseases, it isn’t unusual that many of them remain undiscovered until they are officially curated in an panel-reviewed database such as Online Mendelian Inheritance in Man (OMIM)5 several months later. An automated early-curation system that identifies diseases, symptoms, genes and mutations in these works would reduce the temporal gap between research and clinical application by weeks to months. While individual collections of research may not be as definitive as a fully-curated database entry on OMIM, they are a light at the end of the tunnel for cases where no other references can be found. (Shameless plug: 3billion provides a follow-up of undiagnosed cases when diagnosis becomes possible, at no additional cost.)
Suggesting potentially pathogenic variants with limited traditional evidence
The current gold standard for genetic disease diagnosis is the joint consensus recommendations regarding variant interpretation set forth by the American College of Medical Genetics and the Association for Molecular Pathology6 (henceforth “The Guidelines”). The Guidelines allow for the use of computational predictions as supporting evidence in the determination of variant pathogenicity. An AI trained to suggest potentially pathogenic variants could help diagnose borderline cases when not enough traditional indications are available.
Conclusion
The intersection of AI and rare disease diagnosis holds great potential; even relatively minor features offer tangible benefits in time and diagnosis rate. Other possibilities not listed here also exist, but greater levels of AI involvement also raises the amount of proof expected in its evaluation. It’s not to say that AI algorithms will never be more than a supporting role-- but until the elephants in the room, the requirements of reliability, of explainability, and difficult issues regarding outcome responsibility are addressed, applications that make more significant decisions in the clinical process will likely continue to face headwinds not only at the regulatory level but also at the healthcare institution and patient levels.
Trust is built through transparency and continued validation. Though continued validation is already possible, methods to clarify the inner workings of AI models are still being researched in the fields of explainable AI (why did the model say that?) and uncertainty quantification (teaching the model to say “I don’t know”). The role of AI in rare disease diagnosis is expected to expand as foundations for its credibility further strengthen.
References
- Greenspan A. California DMV Tesla Robo-Taxi / FSD E-Mails. https://www.plainsite.org/documents/242a2g/california-dmv-tesla-robotaxi--fsd-emails/. Page 16 in document.
- SAE International. SAE International Releases Updated Visual Chart for Its “Levels of Driving Automation” Standard for Self-Driving Vehicles
- Monarch Initiative. Human Phenotype Ontology
- 3billion. Internal data on the relationship between the number of phenotype annotations and diagnosis rate. Accessed December 20, 2021
- Johns Hopkins University. Online Mendelian Inheritance in Man
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and genomics and the Association for Molecular Pathology. Genetics in Medicine. ;17(5):405-424. doi:10.1038/gim.2015.30 (2015).