Types of Pathogenic Variants in Rare Diseases and Their Diagnosis
What do you know about rare genetic diseases and how they are diagnosed?
Understanding the Genetic Underpinnings of Rare Diseases
Understanding the genetic basis of rare diseases is a complex but necessary task. The challenge begins with the diverse nature of pathogenic variants, which are pivotal in causing disease but vary significantly in their structure and impact. Diagnosing rare diseases through genetic means is akin to solving a puzzle where identifying the exact cause can be immensely difficult.
The Diversity of Pathogenic Variants
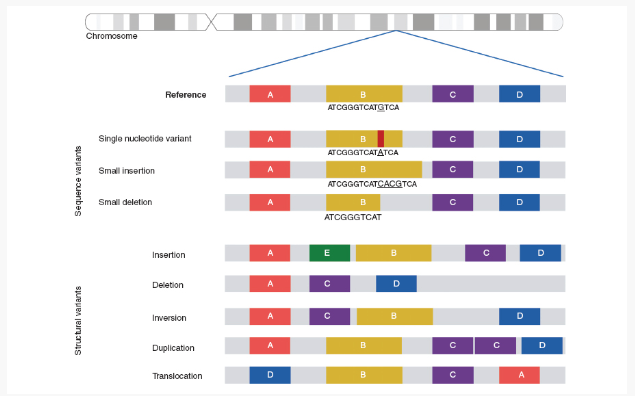
1. Small-Scale Variants: SNVs and Indels
Single Nucleotide Variants (SNVs) and small insertions or deletions (indels) are common genetic changes at the DNA level. Despite their small size, these variants can have a profound impact by altering protein function, leading to disease. For example, a specific SNV in the IRGM gene has been linked to Crohn’s disease, impacting gene expression and contributing to disease pathogenesis. Identifying such variants is crucial in understanding the genetic basis of rare diseases.
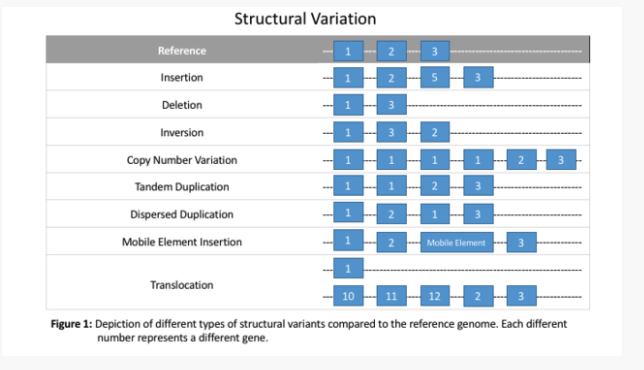
2. Structural Variants: Larger-Scale Changes
Structural variants, such as duplications, deletions, inversions, and translocations, represent large-scale changes in DNA. These variants can disrupt entire genes or regulatory elements, leading to complex disease mechanisms. For instance, the FTO gene locus associated with obesity impacts the expression of the IRX3 gene, significantly influencing body weight and metabolism. Identifying structural variants is more challenging than smaller mutations due to their size and complexity, but their role in rare diseases is vital to uncovering genetic causes.
3. Noncoding Variants: Beyond the Protein-Coding Regions
Noncoding variants occur outside the protein-coding regions of the genome but play crucial roles in regulating gene expression. These variants can disrupt regulatory elements, such as enhancers or promoters, and lead to inappropriate gene activity. Research in fields like epigenetics and single-cell genomics is expanding our understanding of how noncoding variants contribute to disease, offering insights into their functional roles in different cell types and tissues.
The Nature of Mutations: Loss vs. Gain of Function
The nature of a mutation—whether it’s a loss-of-function variant that reduces protein activity or a gain-of-function variant that results in increased or abnormal activity—adds another layer of complexity to genetic diagnosis. Each type of mutation presents unique challenges in detection and interpretation, further complicating the search for the underlying causes of rare diseases.
Diagnosing Rare Diseases: Methods and Challenges
Diagnostic Methods
• Whole Genome Sequencing (WGS): WGS analyzes the entire genome, including both coding and noncoding regions. It is particularly useful in uncovering novel or rare variants that contribute to rare diseases.
• Whole Exome Sequencing (WES): WES focuses on the exome, the protein-coding regions of the genome. It is cost-effective and widely used to identify variants in known disease-causing genes.
• Targeted Sequencing: This approach involves sequencing specific regions of the genome, often when a particular variant or set of variants is suspected to be involved in a disease.
• Single-Cell Genomics: This cutting-edge technique allows for the examination of genetic variation at the single-cell level, offering detailed insights into how variations in individual cells contribute to disease.
• Bioinformatics Tools: Advanced software and algorithms are used to analyze sequencing data, predict the impact of genetic variants, and determine their relevance to disease.
Challenges in Genetic Diagnosis
• Interpretation of Variant Significance: Determining whether a variant is pathogenic, benign, or of uncertain significance remains a challenge. This requires understanding the variant’s impact on protein function and its prevalence in the general population.
• Ethnic and Population Diversity: Genetic variation differs across populations, and the lack of representation of diverse ethnic groups in genetic databases can limit accurate identification and interpretation of variants in underrepresented populations.
• Technological Limitations: While sequencing technologies have advanced, detecting certain types of variants, such as structural variants or those in repetitive genomic regions, remains difficult.
• Data Analysis and Storage: The large volume of data generated by sequencing requires robust computational tools and significant storage capacity for effective analysis.

Claim Your Free Copy of the 2024 Rare Disease Report!
Explore breakthroughs and solutions shaping rare disease diagnostics
• Clinical Translation: Applying genetic findings to clinical practice is an ongoing challenge, particularly when integrating genetic data into patient care and using it to guide treatment decisions.
The Need for Advanced Diagnostic Tools
As medical professionals, it is essential to continuously adapt to the evolving landscape of genetic testing. Tools like chromosomal microarray analysis (CMA) and targeted panel sequencing have been invaluable, but with the advent of technologies like Whole Genome Sequencing (WGS) and Whole Exome Sequencing (WES), the potential for diagnosing rare diseases is expanding.
WGS and WES offer a more comprehensive view of the genome, revealing insights that traditional methods might miss, such as structural variants and changes in noncoding regions. These tools are particularly important for rare diseases, where every genetic clue matters. Additionally, advancements in bioinformatics have improved our ability to interpret complex genomic data with greater accuracy and efficiency.
As we transition to these advanced methods, the potential for improving patient care is immense. By integrating WGS and WES into standard practice, we can enhance our ability to diagnose and treat rare diseases, ultimately improving outcomes for patients and families seeking answers.ust for rare diseases but for a broader spectrum of conditions. The question is, are we ready to transition and integrate these powerful tools into our practice for the betterment of patient care?
References
Get exclusive rare disease updates
from 3billion.

Sree Ramya Gunukula
Marketing Leader with experience in the pharma and healthcare sectors, specializing in digital health, genetic testing, and rare disease diagnostics.







