Sample preparation guide for WES provided by a quality professional
In a previous post, we examined how QC is performed for a WES test for rare disease diagnosis. If QC fails, sequencing cannot proceed as usual, so 3billion will request another sample. In this case, it inevitably takes 2-3 more weeks and more money due to additional sample collection and shipping, so it could feel like a waste of time for the requester of the test.
When a DNA sample is sent for sequencing, the main cause of QC failure is often insufficient sample concentration. Let’s see how best to prepare a sample to avoid this.
- The quality of DNA samples recommended by 3billion is as follows.
- Total amount: 2.0 μg of intact gDNA
- Concentration: 20 ng/ul
- Volume: 100 μL
- Purity: A260/A280 > 1.7
- Buffer: TE 1
Though a spare amount is requested in case of reruns, if a sample is sent according to an inaccurate measurement of DNA concentration, QC may fail due to an insufficient amount of DNA. The cause of inaccurate measurement of DNA concentration may be due to the experimenter, but it mostly depends on the measuring equipment. Then, let’s look at the characteristics and differences between Nanodrop and Qubit, which are equipment used mainly to measure DNA concentration.
Difference between Nanodrop and Qubit
- Nanodrop

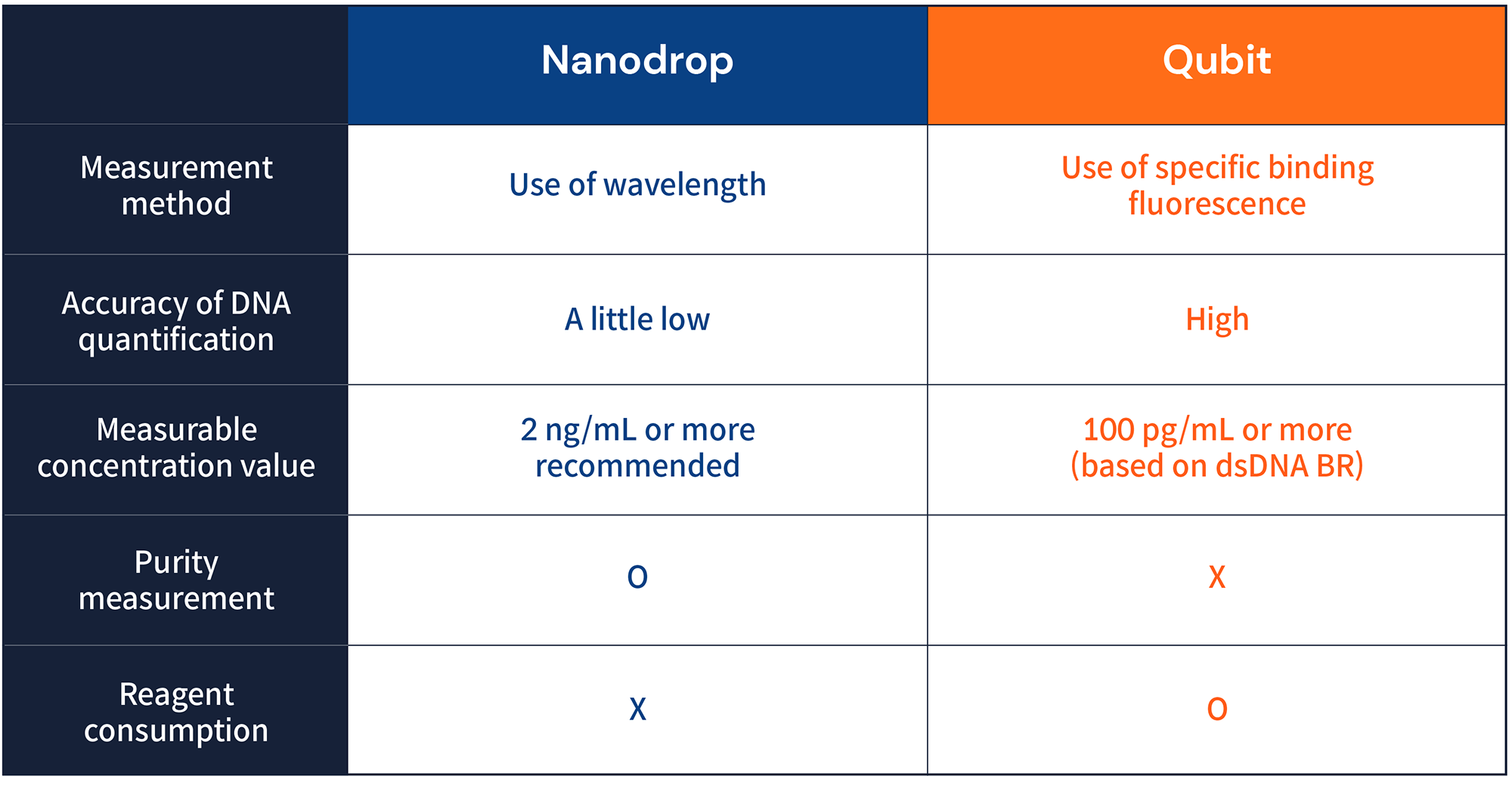
All nucleic acids absorb light with a maximum wavelength of 260 nm, and Nanodrop uses this to measure the concentration of nucleic acids. However, since DNA and RNA are measured without distinction of single strand and double strand, inaccurate concentration values can result, which are also affected by contaminants or changes in base composition. In addition, measurement of samples with a concentration of less than 2 ng/mL is not recommended.2
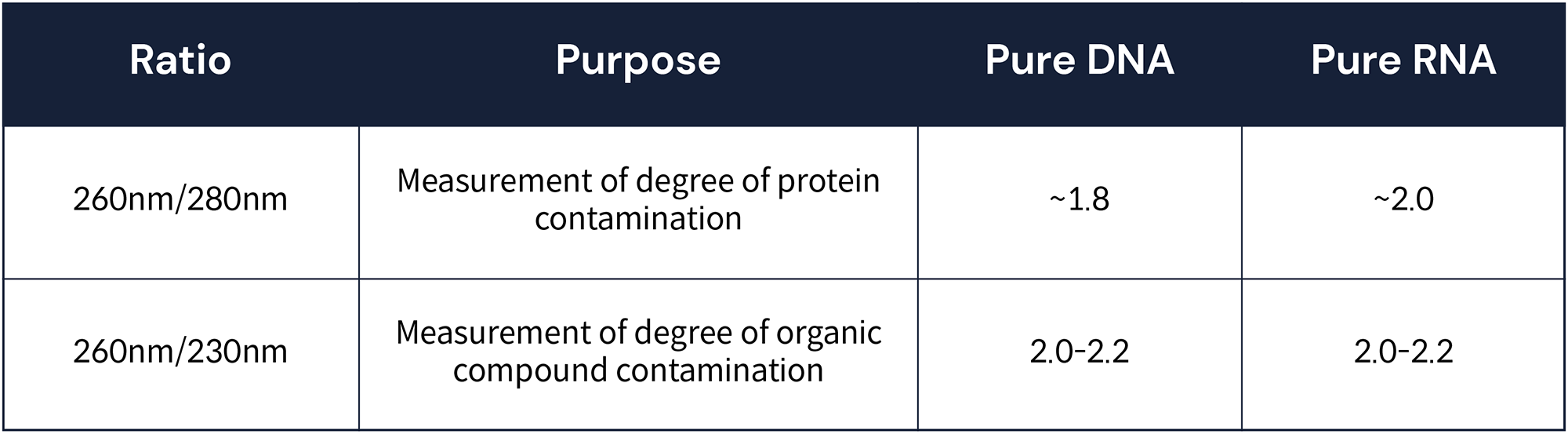
The advantage of Nanodrop is that it does not require any reagents for sample measurement, so there is no cost to measure and purity can be checked. The degree of contamination of organic substances in the extracted sample can be checked with the wavelength of 230 nm, and the degree of contamination with salt and protein can be checked with the wavelength 280 nm. The closer the 260nm/280nm value is to 1.8, the purer the DNA, and the closer to 2.0, the more likely it is pure RNA. Also, when the 260nm/230nm value is 2.0~2.2, it is pure DNA or RNA.3
- Qubit

Qubit is a fluorescence-based measuring device, and it is possible to obtain a more accurate concentration value without being affected by contaminants by binding to a specific nucleic acid to be measured. In addition, it can detect even the smallest concentrations (100 pg/mL based on dsDNA BR). Disadvantages include that purity cannot be measured and that reagents are required to measure the concentration, which incurs a cost per sample.
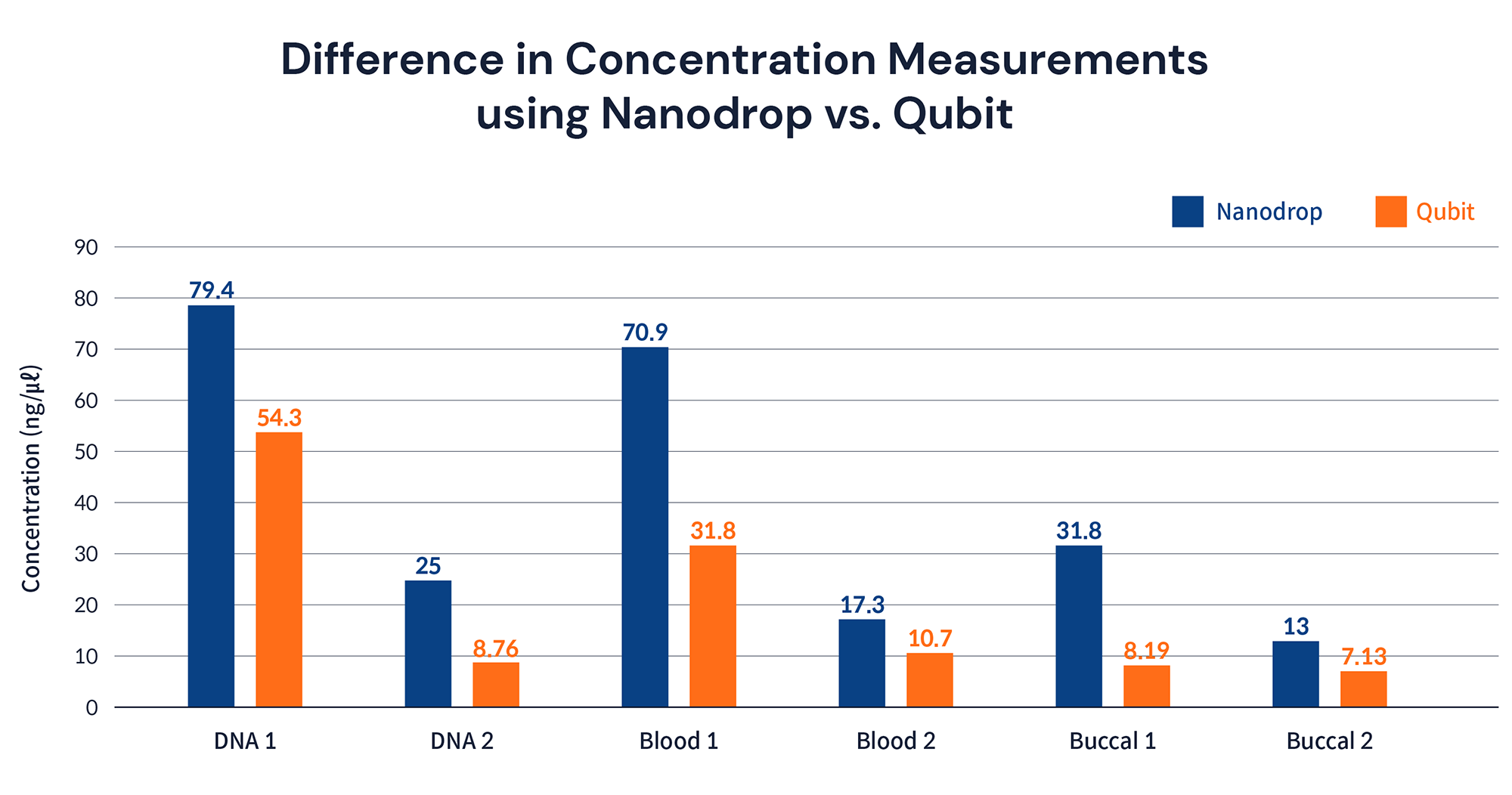
So, how much DNA sample should I prepare according to the device I use?
If the amount of nucleic acid is measured only with Nanodrop, how much more should the sample be compared to measuring with Qubit? As can be seen in the table below, it is recommended to prepare 1.5~2 times the required amount, though the ratio value is different depending on the degree of contamination of each sample.
So far, we have looked at QC issues that may appear when sending DNA samples for WES and ways to prevent them.
Sample preparation is important, but in many cases, samples are sent to other regions or other countries because there are no medical institutions with clinical labs locally, and you have to pay attention to storage and transportation of samples. Therefore, I would like to share that the best way to obtain WES analysis results is to send the sample in compliance with the following storage methods.
- Blood
Cells with nuclei in the blood are white blood cells. The amount of DNA extracted depends on the viability of the white blood cells. Because the survival rate of leukocytes in whole blood decreases at 4°C and -20°C, the DNA yield decreases by 30-40%. This is far less than the amount of DNA that can be extracted from fresh blood, but quality is not affected. Therefore, DNA extraction should be done as quickly as possible, and if extraction is not possible immediately, the sample should be stored at 4℃ or -20℃.4 It is best to store it at room temperature for less than 2 days, and when shipping after refrigeration or freezing, pack it in an ice box with ice packs. Only whole blood in an EDTA tube can be accepted at 3billion, and it is recommended to provide 1mL or more.
- Buccal Swab
Buccal swabs are convenient because samples can be collected non-invasively. With the two provided cotton swabs, scrape the inside of the cheeks at least 20 times each and put it in the container with the buffer. Samples that have been collected should be stored at room temperature and sealed so that the buffer does not leak. If stored for a long period of time, bacteria may proliferate, so it is recommended to ship it within 1 week.5
- DNA
In order to preserve extracted DNA for a long time, it is recommended to use a Tris-EDTA (TE) buffer to extract it and keep it refrigerated at 4℃. For long-term storage, it is recommended to store at -20℃ or -70℃.6 Repeated freezing and thawing can cause DNA shearing, so caution must be taken when freezing samples. When shipping, the DNA sample should be sealed so that it does not dry, and it can be shipped at room temperature for same-day delivery. Otherwise, it is recommended to enclose ice packs in the shipment.

Discover Our Advanced Whole Genome Sequencing!
Get the most detailed and accurate Genome Test available.
References
- invitrogen, Comparison of fluorescence-based quantitation with UV absorbance measurements, https://tools.thermofisher.com/content/sfs/brochures/fluorescence-UV-quantitation-comparison-tech-note.pdf
- GW Vitek, http://www.afrontier.net/laboratory/product/item2.php?it_id=1586324500
- Salman A.H. AlRokayan, 「Effect of Storage Temperature on the Quality and Quantity of DNA Extracted from Blood」, Pakistan Journal of Biological Sciences, 3(3), 2000, p392-394
- 3billion, https://3billion.io/blog/how-do-you-decide-between-blood-or-buccal-swab-collection/
- Yun-Tae Kim et al, 「Effects of Storage Buffer and Temperature on the Integrity of Human DNA」, Korean J Clin Lab Sci, 44(1), 2011, p24-30
Get exclusive rare disease updates
from 3billion.








