Genomic Newborn Screening (gNBS): Quickly Identifying and Addressing Your Child’s Genetic Risks!
Recently, research into genomic Newborn Sequencing (gNBS) has been actively progressing worldwide. This innovative approach offers the opportunity for early diagnosis and treatment of potential genetic disorders in apparently healthy newborns. This article, based on a recent paper, delves into what gNBS is, its current global status, and the future directions it should pursue.
What is gNBS and What is its Purpose?
gNBS is the process of early detection of the risk for a wide range of genetic disorders through newborn and childhood sequencing (NBSeq).
Purpose:
- To discover the risk of genetic disorders early in apparently healthy infants.
- The aim is to prevent irreversible damage and improve treatment outcomes through early diagnosis and intervention.
- Interest in gNBS is growing as over 700 genetic disorders now have targeted treatments or consensus guidelines for long-term management.
Global Status of gNBS Programs: Who and Where?
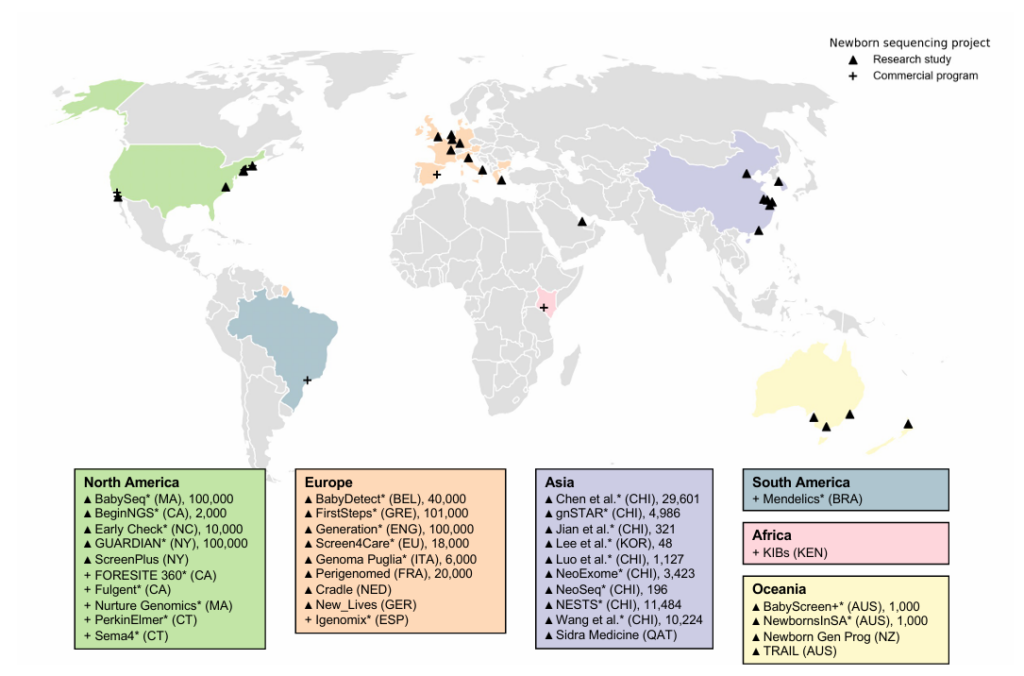
- Currently, over 30 international research programs and commercial companies are actively exploring the gNBS approach. Many of these programs exchange best practices through the International Consortium on Newborn Sequencing (ICONS).
- A total of 35 research and commercial programs were identified. Of these, 10 were in North America, 10 in Asia, and nine in Europe.
- Results from nine research programs covering a collective total of 68,628 infants (up to September 2024) showed the percentage of positive screening results ranged from 1.85% to 9.43%, with an average of 3.82%.
출처: Genet Med. 2025 Jul;27(7)
Comparison and Differences in Gene Lists Across Programs
An analysis of 4,390 genes included by 27 gNBS programs revealed substantial heterogeneity across the gene lists.
🔍 Key Findings
- Gene List Size: The number of genes analyzed ranged widely from 134 to 4,299 (median = 306).
- High Concordance Genes: Out of 4,390 genes, only 74 genes (1.7%) were included by over 80% (22 of 27) of the programs.
- Major Clinical Areas: Most genes were linked to inherited metabolic disorders (IMDs, 25.4%), followed by neurologic (15.5%) and immunologic (11.9%) disorders.
- US RUSP Influence: The presence on the US Recommended Uniform Screening Panel (RUSP) was the most significant factor associated with gene inclusion.
Factors Predicting Gene List Inclusion
Regression analysis identified key characteristics strongly associated with inclusion across programs.
Strongest Predictors:
- RUSP Inclusion: Core conditions showed an inclusion increase of 74.7% , and secondary conditions, 60.0%.
- Robust Evidence Base (Natural History): 29.5% increase in likelihood of inclusion.
- High Treatment Efficacy: 17.0% increase.
- Expert Recommendation Score: Genes recommended by ≥80% of experts were 43.5% more likely to be included.
Other Important Predictors: High Penetrance , Neonatal or Infantile Onset , high Disease Severity , and Treatment Acceptability.
The Future of gNBS: What Direction Should We Take?
The substantial variation in gene lists highlights the need for a consistent and evidence-based method for gene selection.
Data-Driven Approach (Machine Learning)
- The study developed a boosted trees machine learning model to provide a data-driven approach to gene selection.
- Model Features: The model uses 13 gene-disorder characteristics (e.g., RUSP category, clinical area, treatment efficacy) to predict gene inclusion, achieving high accuracy (AUC=0.915, R2=84%).
- Utilization: This model generates a dynamic ranked list of genes that can adapt to emerging evidence and regional needs, helping public health programs prioritize genes for NBSeq implementation.
- Identifying Overlooked Genes: The model can identify highly ranked genes (e.g., PTPRC, CYBA) that were only included by a few current programs, potentially identifying genes overlooked by researchers.
Future Challenges and Improvement Directions
- Data Enhancement: Future research plans include integrating perspectives from key informants like parents and pediatricians and incorporating additional bioinformatics data (e.g., gene length, gnomAD constraint score).
- Knowledge Expansion: Researchers plan to use a knowledge graph to identify genes that are not currently included but share similar characteristics (e.g., common molecular pathways or treatments) with highly represented genes.
- Flexibility and Consistency: The goal is to move beyond a static list to a dynamic ranking system that can be updated as new knowledge and therapeutics emerge, contributing to international consensus and consistency in gNBS gene selection.
gNBS is a crucial turning point for the early detection of genetic disorders. In selecting genes, there is no single “correct” answer: some programs may prioritize Sensitivity (maximum risk detection), while others focus on Specificity and “Clinical Utility” (only highly actionable conditions). The data-driven approach, accumulating information from diverse projects, is expected to play a key role in building a trustworthy screening strategy.
We hope this post enhances your understanding of gNBS and provides useful material for future discussion.
Get exclusive rare disease updates
from 3billion.

Sookjin Lee
Expert in integrating cutting-edge genomic healthcare technologies with market needs. With 15+ years of experience, driving impactful changes in global healthcare.