Which is the Best NGS Approach for Rare Disease Diagnosis: Panels, WES, or WGS?
As next-generation sequencing (NGS) costs and analysis times continue to decrease, the technology has expanded its role in diagnosing rare genetic diseases. Annually, 250 gene-disease associations and > 9,000 variant-disease associations are reported, which continuously increase our understanding regarding genomes.
While NGS panels are still widely used in clinical settings, the adoption of whole exome sequencing (WES) and whole genome sequencing (WGS) is steadily increasing. However, choosing the best NGS testing method depends on the patient’s unique case.
This article outlines the characteristics, limitations, and recommended uses of NGS panels, WES, and WGS, helping clinicians select the best approach.
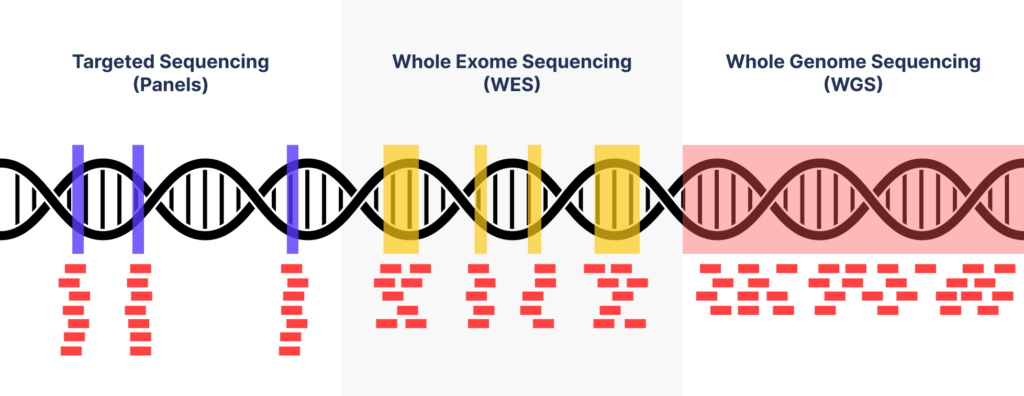
Panels: Targeted Sequencing
Panels, targeted sequencing represents a method of selecting and analyzing genes that are related to a specific disease or disease group.
It is the most economical and effective diagnostic approach in cases wherein the genes of the suspected diseases have already been identified. It has greater coverage and higher depth per base of targeted genes, so it’s easier to interpret the report results.
Owing to the low initial diagnostic rates of rare diseases, reanalysis and reclassification is important. Like some studies have demonstrated, important amounts of rare disease diagnosis encompasses recent-known diseases: among positive WES findings, 23% are within genes discovered within the last two years and 7% of them are novel gene discoveries.
However, in most cases, targeted sequencing affords only a one-time anaylsis. It means, if the prescribed panel result is negative, it is impossible to re-analyze the other panel.
Whole Exome Sequencing (WES)
Whole exome sequencing focuses on the exon regions, which comprise only 2% of the genome but hold 85% of known pathogenic variants. WES is typically more cost-effective than WGS and provides more extensive information than targeted sequencing. It’s an ideal first-tier test for cases involving severe, nonspecific symptoms or conditions like chromosomal imbalances, microdeletions, or microduplications
However, WES does have certain limitations. Firstly, not all exonic regions can be evaluated, and noncoding regions are not sequenced, making it impossible to detect functional variants outside the exonic areas. Additionally, except for a few cases of copy number variations (CNVs), such as indels and duplications, WES shows low sensitivity to structural variants (SVs).
Moreover, the results of WES can vary depending on the entity being analyzed, even when the same WES method is used. The targeted regions and probe manufacturing methods differ based on the test kit used, which can lead to variations in data quality. On average, approximately 100,000 mutations can be identified in an individual’s WES data, and these mutations are interpreted according to the guidelines of the American College of Genetics and Genomics (ACMG), which prioritize those most closely related to the patient’s symptoms.
Due to these limitations, additional testing may sometimes be required. Nevertheless, because WES requires fewer sequencing reagents compared to WGS, it remains recommended as a first-tier test for certain cases, particularly those involving severe symptoms during the neonatal period or childhood, cases with extensive, complex, and nonspecific symptoms, suspected chromosomal imbalances, microdeletions or microduplications, and pathogenic mutations that could not be detected by previous genetic tests.
Whole Genome Sequencing (WGS)
WGS involves analysis of the entire genome; hence, it’s diagnostic rate is the highest among all the genetic testing methods. In other words, the application of this method as a first-tier analysis will extend gene analysis coverage to noncoding regions, thereby reducing unnecessary repetitive testing. Further, this method allows the detection of variations, such as CV and mitochondrial DNA, which cannot be valuated using WES.
However, WGS has certain limitations as well. Compared with WES, WGS generates extensive data, and the cost of storing and analyzing this data is two to three times higher than that of WES, although constant technological advances are steadily leading to a decrease in these costs. In addition, there is insufficient research on noncoding regions compared with exonic regions, resulting in inadequate information for variant analysis; this insufficient evidence regarding pathogenicity can lead to confusion among clinical geneticists.
BEST NGS Approach
On average, the diagnostic odyssey for patients with rare genetic diseases spans 7 years. Given that 80% of rare diseases have a genetic basis, the advancement of next-generation sequencing (NGS) is crucial for faster diagnosis. However, while whole genome sequencing (WGS) offers comprehensive coverage, its ability to predict certain rare diseases remains challenging due to the wide variety of symptoms involved. This may improve with further technological advancements.
Currently, WGS is recommended only when pathogenic variants are not detected through targeted sequencing or whole exome sequencing (WES). On average, WGS detects around 3 million mutations, making it nearly impossible to assess the pathogenicity of each mutation individually. To address this, several research groups, including 3billion, are developing AI algorithms and technologies to narrow this gap. The 3billion AI model 3Cnet has demonstrated high accuracy in predicting variant pathogenicity using clinical data from ClinVar and topped the 6th Genome Analysis Critical Assessment.
To choose the most suitable NGS approach for each patient, clinicians need to consider various factors, including the diagnostic stage, symptoms, patient age, medical and family history, as well as the economic situation. Guidelines based on reliable public sources, newly published studies, and clinical experience should guide the selection of the appropriate genetic testing method.
In 2021, the American College of Medical Genetics and Genomics (ACMG) recommended both WES and WGS as primary or secondary testing options for patients with rare genetic diseases, such as congenital abnormalities, developmental delays, or intellectual disabilities (CA/DD/ID). Numerous studies have shown that WES and WGS can significantly increase diagnostic rates and provide greater clinical utility in such cases. At 3billion, we hope that our 3B-GENOME and 3B-EXOME services can further aid in the diagnosis of rare diseases for many patients.
Understanding the Limits of WES
While WES is often a preferred option for rare disease diagnostics, it’s important to understand where it might fall short. Knowing its limitations can help set realistic expectations and guide more informed decisions—especially when diagnostic answers remain elusive.

Still Unsure Which Test Fits Your Needs?
Schedule a free consultation with our genomics expert to choose the best option for your patient.
Get exclusive rare disease updates
from 3billion.

Sree Ramya Gunukula
Marketing Leader with experience in the pharma and healthcare sectors, specializing in digital health, genetic testing, and rare disease diagnostics.







