Rare Disease Diagnosis in the Middle East: How WES and WGS Are Improving Outcomes
Rare diseases affect approximately 300 to 400 million people globally, with an estimated prevalence of 6% to 8% of the population. In the Middle East and North Africa (MENA) region, the burden of rare diseases is significantly higher due to several socio-demographic and genetic factors, including high rates of consanguinity, large family sizes, and limited access to advanced diagnostic tools. Despite the increasing awareness and medical advancements, the diagnosis and management of rare diseases in the MENA region remain fragmented and underdeveloped.
A comprehensive approach to profiling rare diseases in the MENA region is critical for improving early diagnosis, personalizing treatment, and enhancing long-term patient outcomes.
The Landscape of Rare Diseases in the MENA Region
The MENA region presents a unique set of challenges and opportunities when it comes to rare diseases:
1. High Prevalence of Genetic Disorders
- High rates of consanguinity (ranging from 20% to 60% in some populations) increase the risk of autosomal recessive genetic disorders.
- Certain rare diseases such as thalassemia, cystic fibrosis, and metabolic disorders have a higher prevalence in the MENA region compared to Western populations.
- Founder effects and endogamy further contribute to the clustering of rare genetic mutations within specific populations.
2. Underdiagnosis and Misdiagnosis
- Limited availability of genetic testing and specialized healthcare centers leads to delayed or missed diagnoses.
- Misdiagnosis often results in inappropriate treatments, higher healthcare costs, and increased morbidity and mortality.
- The lack of structured data and regional registries makes it difficult to track and study rare disease patterns accurately.
3. Healthcare Infrastructure Challenges
- Limited funding for rare disease research and genetic testing infrastructure.
- Shortage of trained genetic counselors and specialists in rare diseases.
- Inequitable access to specialized care across urban and rural areas.
Comprehensive Profiling Through WES and WGS
Advancements in genomic medicine have opened new avenues for diagnosing and profiling rare diseases. Two key next-generation sequencing (NGS) technologies — Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS) — have proven to be game changers in identifying the genetic basis of rare diseases.
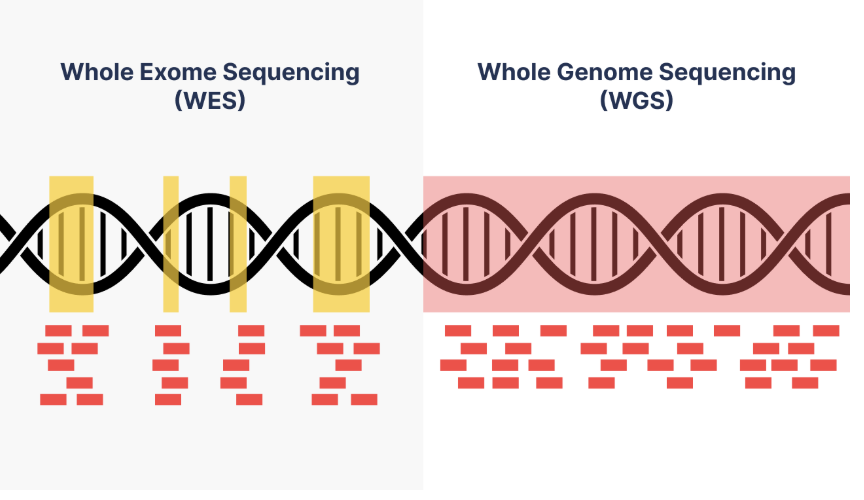
1. Whole Exome Sequencing (WES)
WES focuses on sequencing the protein-coding regions of the genome (exons), which represent approximately 1% to 2% of the entire genome but account for over 85% of known disease-causing mutations.
How WES Can Help in the MENA Region:
- It’s estimated that about 80% of rare diseases are caused by genetic mutations, which means they are inherited or arise from spontaneous genetic changes (de novo mutations). Some examples of hereditary rare diseases include: Cystic fibrosis – Autosomal recessive inheritance, Huntington’s disease – Autosomal dominant inheritance, and Duchenne muscular dystrophy – X-linked inheritance. WES can identify disease-causing mutations in genes linked to rare diseases.
- Cost-effective compared to WGS while still covering the majority of disease-associated variants.
- Particularly effective for diagnosing Mendelian disorders, metabolic diseases, and congenital malformations.
- Useful for targeted diagnostics in populations with high rates of consanguinity. In consanguineous cases, recessive mutations are more likely. WES can identify homozygous mutations (mutations inherited from both parents).
Example:
In a recent study conducted in Saudi Arabia, WES identified pathogenic mutations in over 60% of children with previously undiagnosed neurological disorders, highlighting the utility of WES in populations with high consanguinity.
2. Whole Genome Sequencing (WGS)
WGS sequences the entire genome, including both coding and non-coding regions, providing a comprehensive view of genetic variation.
How WGS Can Help in the MENA Region:
- In consanguineous cases, the genetics tend to be more complex because of increased homozygosity and the higher likelihood of rare recessive mutations. This means the disease-causing mutation might not always be in the coding region. Since WGS covers the whole genome — including intronic and regulatory regions — it can detect mutations that WES would miss. Also, structural variants like large deletions, duplications, and translocations are more likely in consanguineous cases, which WGS is better at identifying.
- Particularly useful for polygenic and complex disorders with unknown genetic etiology.
- Provides data for future research on gene-environment interactions and epigenetic factors.
- Useful in cases where WES yields inconclusive or negative results.
Example:
A study in Qatar used WGS to identify novel pathogenic variants in patients with undiagnosed primary immunodeficiency, allowing for targeted therapies and better clinical outcomes.
How 3billion Enhances Genetic Diagnosis in Consanguineous Cases
1. Enhanced Exome Sequencing
Traditional whole exome sequencing (WES) often misses critical variants in such cases due to insufficient coverage or failure to detect non-coding and mitochondrial mutations. 3billion’s Enhanced Exome (3B-Exome) addresses these challenges through targeted improvements in coverage and detection accuracy.
- Enhanced Coverage of Coding Exons
- Improved Detection of Non-Coding Variants
- Integrated Mitochondrial Genome Analysis
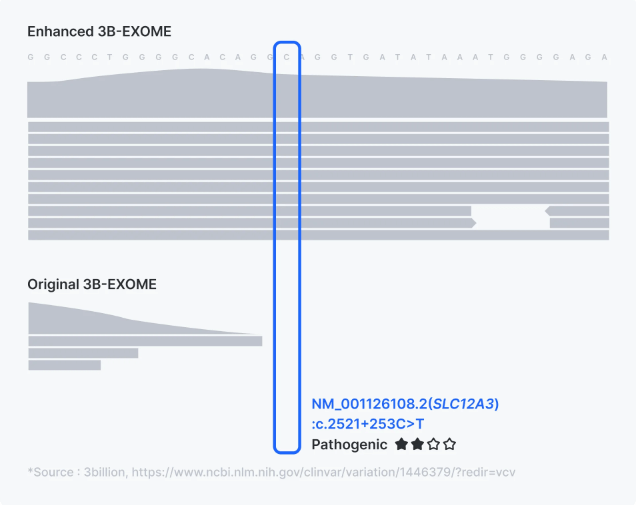
For further details, please refer to the comprehensive article on Enhanced WES.
2. Upgrading to WGS from WES is Critical for Consanguineous Cases
In regions such as middle east where consanguineous marriages are common, the risk of autosomal recessive diseases is significantly higher. Many of these diseases are caused by mutations in non-coding or regulatory regions that standard WES cannot detect.
By enabling a straightforward transition from WES to WGS, 3billion ensures that healthcare providers can adjust their diagnostic approach based on initial findings. This feature ensures more comprehensive analysis without needing to order separate tests, reducing both time and costs

Interested in upgrading to WGS or learning how this feature can benefit consanguineous cases?
Schedule a call with our dedicated team
3. 3B-INTERPRETER for Undiagnosed Cases or Variants of Uncertain Significance (VUS)
For patients who remain undiagnosed after initial genetic testing, 3B-INTERPRETER offers a powerful solution. It leverages AI-driven algorithms to re-evaluate existing WES or WGS data, integrating the latest research and updated clinical information to improve diagnostic accuracy.
3B-INTERPRETER also offers continuous renalysis. It automatically reanalyzes undiagnosed cases daily, incorporating newly published research and evolving clinical insights. This increases the likelihood of identifying pathogenic variants that may have been missed initially.

2024 Rare Disease Diagnostics Report: Cost, Challenges, and Patient Needs
his report reveals the biggest challenges from the patient’s side in 2024.
Get exclusive rare disease updates
from 3billion.

Sree Ramya Gunukula
Marketing Leader with experience in the pharma and healthcare sectors, specializing in digital health, genetic testing, and rare disease diagnostics.






