Reanalysis and reclassification, why are they important?
With the rapid development of genomic medicine and the increased use of WES/WGS tests, it is crucial to note the importance of continuous reanalysis versus one-time data analysis.
Why? Results may change as more information accumulates. Through reanalysis, undiagnosed patients may be diagnosed, or former diagnosis results may become invalid.
What is the difference between reanalysis and reclassification?
Reanalysis is the process of reanalyzing existing genomic data against new information, and reclassification is one of the types of changes in results that may occur through reanalysis.
Types of change in diagnosis through reanalysis
Let’s take a closer look at examples of changes from reanalysis through the paper “Reanalysis of Clinical Exome Sequencing Data” introduced in NEJM in 2019.1 In this paper, change in diagnostic rate and change in results through reanalysis were analyzed in two different cohorts.
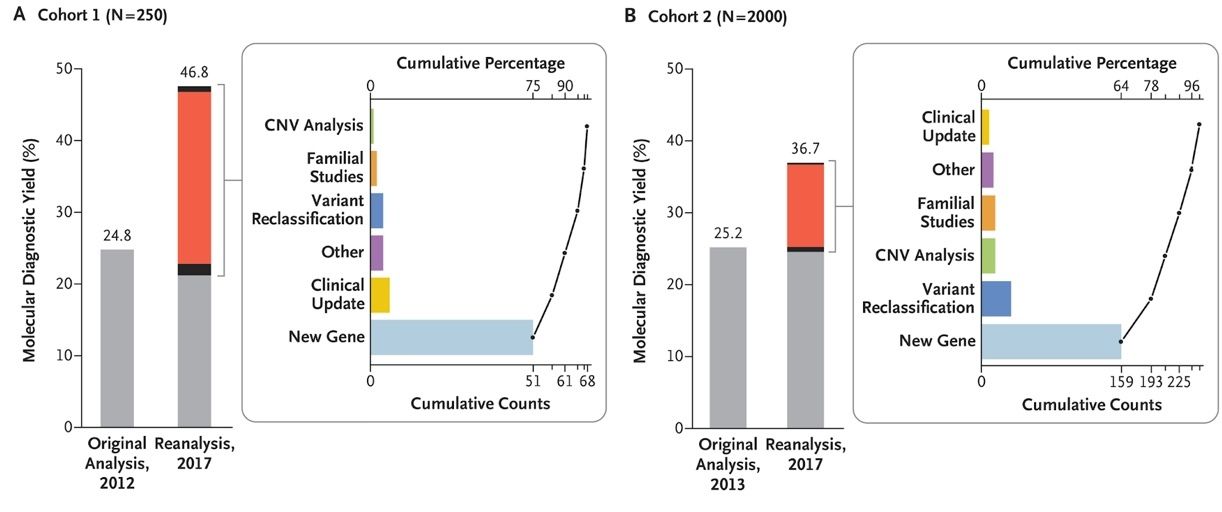
(Reference: https://www.nejm.org/doi/full/10.1056/NEJMc1812033)
- Through reanalysis, additional diagnoses were made in 21.5% and 11.5% of each of the two cohorts, respectively.
- Most cases were as a result of a newly discovered disease-gene association (75% and 64%, respectively).
- 6% and 14% were from reclassifications of known variants, respectively.
- Of the 42 patients newly diagnosed through reanalysis, 17 (40%) underwent a change in treatment and management method according to their new results
As demonstrated through this study, reanalysis results may vary depending on the extent to which new information is incorporated during reanalysis.

Curious How We Boost Diagnostic Accuracy?
Get the newsletter every month and discover the breakthroughs with the latest reanalysis report.
Limitations of reanalysis through reclassification only
Let’s look at the limitations of effectiveness when reanalysis was performed only with the reclassification of select genes in a study2. The data from 104 patients diagnosed with inherited arrhythmogenic syndrome (IAS) and 17 patients whose death was found to be caused by IAS postmortem in 2010 were reanalyzed with the reclassification of 128 variants in 17 genes associated with IAS in 2020.
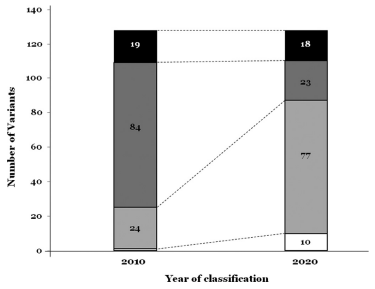
(Reference: https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(20)30107-9/fulltext)
- 71.87% (92/128) variants were reclassified.
- There was no significant change in the Pathogenic (P) variants (19 of 128; 14.84%) that caused the disease after reanalysis (18 of 128; 14.06%).
- Likely pathogenic (LP) variants (84 of 128; 65.62%) were the majority, but after reanalysis, VUS (77 of 128; 60.65%) became the majority.
- Only 23 variants (17.96%) remained as LP variants.
- LB variants increased from 1 to 10 after reanalysis.
Although belonging to the same IAS disease group, the variant reclassification results were different depending on the subtype, and most studies of reanalysis through reclassification demonstrate that VUS is more often downgraded to LB/B than upgraded to P/LP. This can be interpreted that as data of various ethnicities have accumulated, variants that were previously considered rare are no longer considered rare.
As demonstrated by research, diagnosis through reanalysis is mostly as a result of the discovery of new disease genes. Therefore, it is important not only to reclassify known genes and their variants, but also to reanalyze the entire genetic region with any changes in symptoms on a regular basis (at least once a year).3 The search for a diagnosis should be continued relentlessly until diagnosis is confirmed against symptoms and the appropriate treatment is administered.
Have you tried genetic testing in the past but didn’t get a diagnosis? 3billion will help solve the answer with you.
References
- Liu et. al., Reanalysis of Clinical Exome Sequencing Data. N Engl J Med 380, 2478 (2019).
- Campuzano et.al., Reanalysis and reclassification of rare genetic variants associated with inherited arrhythmogenic syndromes. EBioMedicine 54, 102732 (2020).
- Ji et. al., Clinical Exome Reanalysis: Current Practice and Beyond. Molecular Diagnosis & Therapy 25, 529 (2021).
Get exclusive rare disease updates
from 3billion.

Sookjin Lee
Expert in integrating cutting-edge genomic healthcare technologies with market needs. With 15+ years of experience, driving impactful changes in global healthcare.







