Practical tips on understanding whole exome sequencing test results

Whether it is a WES/WGS test for clinical purposes or a DTC test from 23andme or Ancestry that provides ancestry information, a genetic test includes a final results report on what was tested. WES/WGS testing for rare disease diagnosis covers the entire exome or genome and serves the purpose of clinical diagnosis, which in turn can make the report seem complicated and difficult to interpret. But is it, really?
Since WES/WGS tests for clinical purposes are performed in compliance with ACMG standards and guidelines, reports may be very similar even if the testing is conducted in different laboratories.
Let’s take a look at a sample report for 3billion’s rare genetic disease test and break down the different parts of the report.
How is the WES test report organized for clinical purposes?
A WES test report can generally be divided into two sections: the test result section and the additional information section.
The test result section contains order information, test result, and result interpretation, while the additional information section contains secondary findings, target region information, test methods, and limitations.
Test result section

The test result section(p.1-2) of a positive 3billion test report
The test result section displays important results of the test and is generally divided as follows:
Order information
This part includes the ordering physician’s information, institution information, and patient information. Patient information includes the test type, sample type and collection date. Test type refers to whether the patient’s sample is being tested alone or in combination with samples from one or two parents.
Test result
This is the most important part of the report. Genetic test results are shown as “Positive,” “Inconclusive,” or “Negative,” and the accompanying details below vary depending on the result.
- Positive
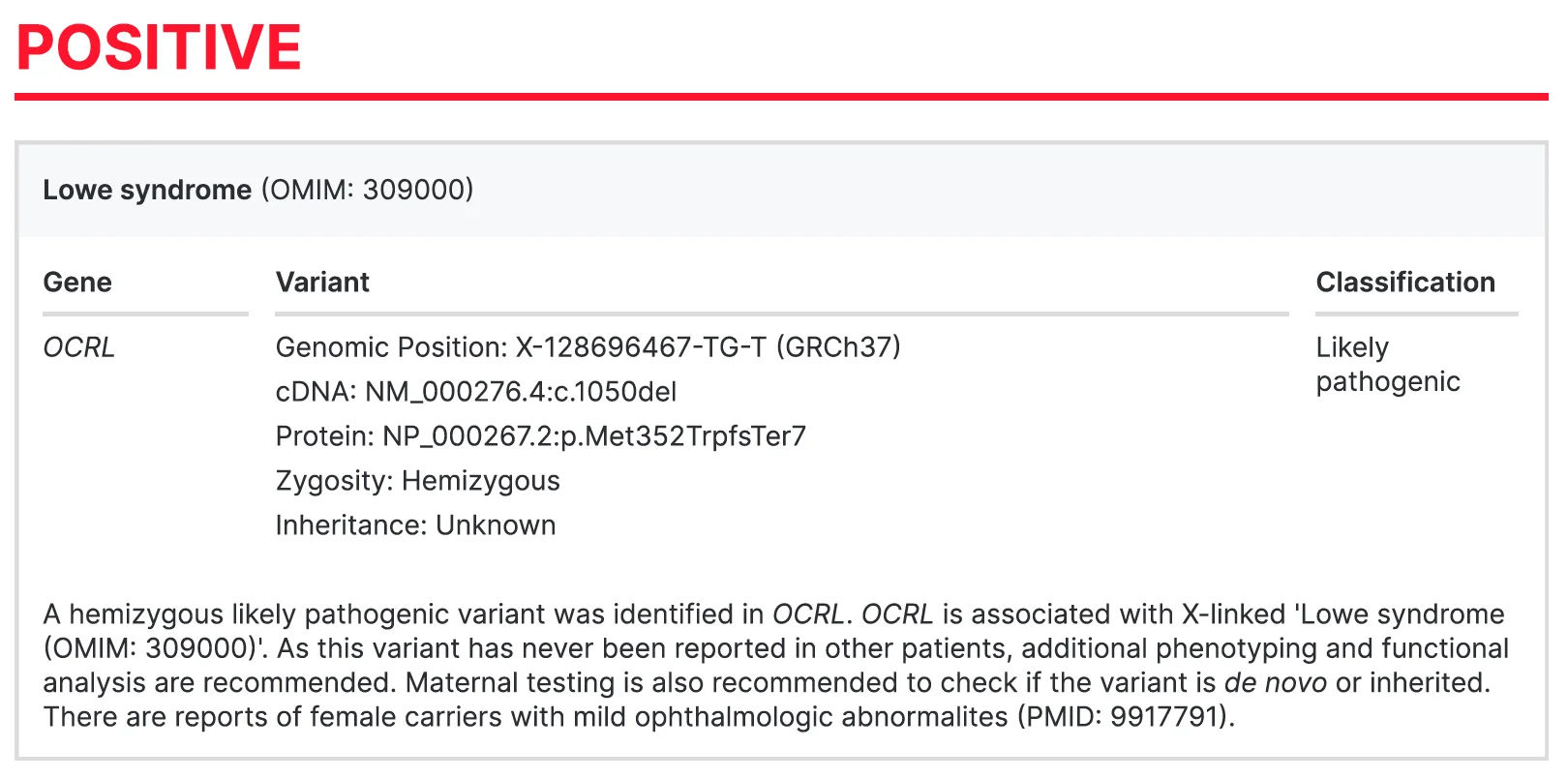
A positive result suggests that the cause of the patient’s disease has been identified through the test. One or more genetic variants that have a 90% or higher likelihood of causing the disease are reported.
- Inconclusive
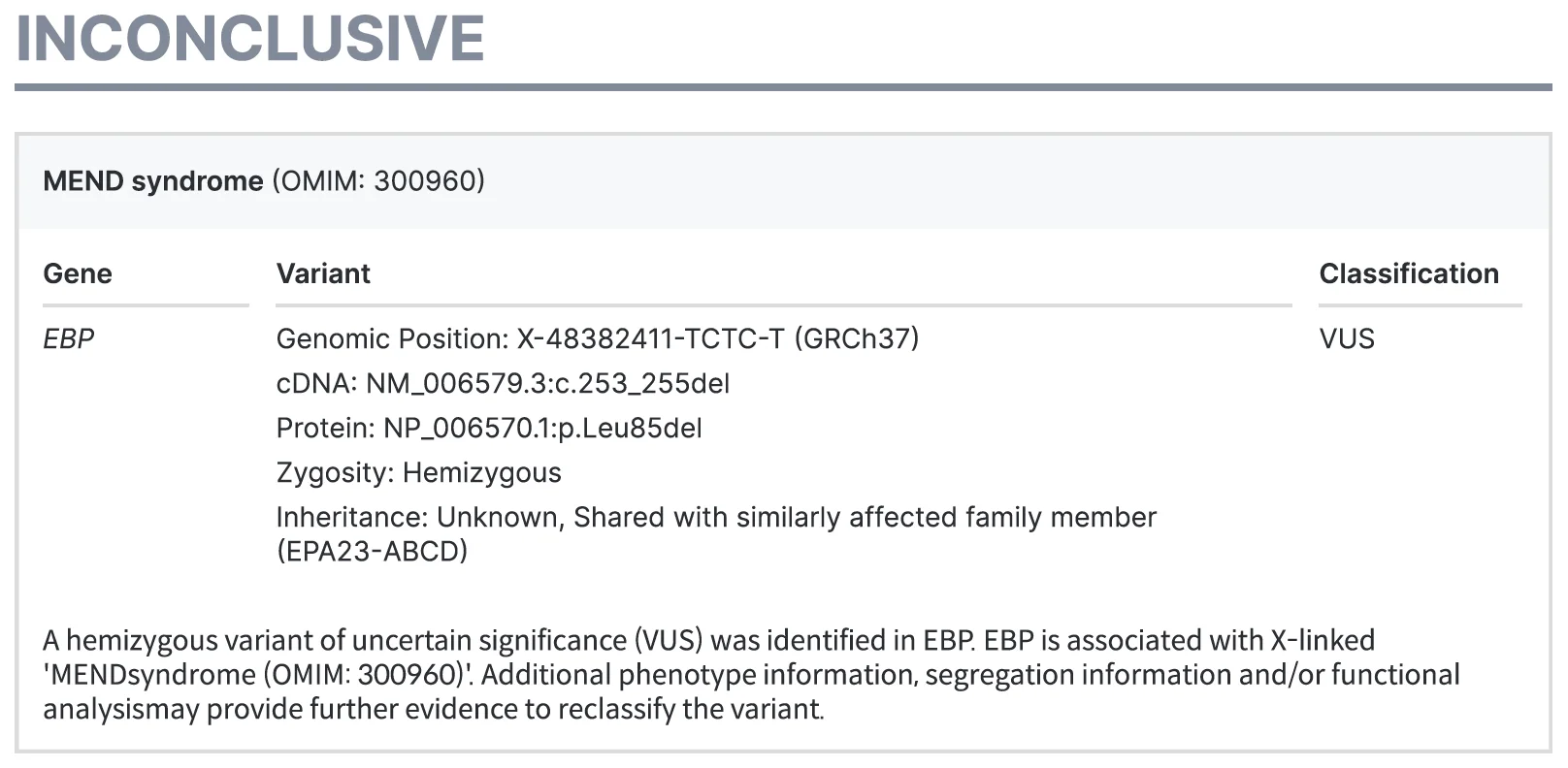
An inconclusive result indicates that the likely cause of the disease has been identified through the test, but the current supporting evidence is insufficient for an accurate diagnosis to be made. Suspicious variants are reported, along with variant classification information and the supplemental information needed to reclassify variants.
- Negative

A negative result indicates that no variant that may have caused the patient’s symptoms was identified.
Result interpretation details
This is the section where you can see the basis for result interpretation in more detail. Although the names may be slightly different for each WES test, they are usually called ‘result interpretation’, ‘results and interpretations’, or ‘variant interpretation’, etc.
- Positive
For positive results, this section provides the evidence used to identify a specific variant as the causative variant for diagnosis. Depending on the laboratory, the format may vary from simple text to a table, but the section mainly details the evidence that supports why the variant has been identified as causative.
In addition, detailed information about the diagnosed disease is also provided. The section usually includes information on the genes and variants associated with the disease, the inheritance pattern of the disease, and the symptoms it presents.
- Inconclusive
For inconclusive results, this section provides detailed information on evidence that classifies the variant as a VUS. The information needed to provide positive/negative results can be found in the “Test result” section.
- Negative
For negative results, this section reiterates that a causative variant was not identified by WES. Since a diagnosis could not be made, it often provides reasons a causative mutation was not found. The most prevalent reason includes the technical limitations of current sequencing and analysis technology, and the limitations of the genotype-phenotype association information known to date.
A negative report can cause disappointment or confusion to both doctors and patients. However, if doctors share with their patients that new rare disease information is constantly being discovered and that technologies to overcome the current limitations are continuously being developed, it will be comforting to know that the possibility of finding answers in the future is always open.1
Additional information section
The main test results are delivered in the first part of the report, followed by supplemental content as described below.
Secondary findings
These are the analysis results of 73 genes recommended by ACMG that WES/WGS patients can receive. Results are usually included by the patient’s choice.
If you would like to read more details on what secondary findings are, why ACMG recommends it, and what to consider when deciding, please refer to 3billion blog’s post on secondary findings.
Target region information
This section mentions the sequencing quality of the sample. Generally, the target region coverage is shown in a table by mean depth and sequencing depth. Depth refers to how many times each DNA region was sequenced and read repeatedly, and it is expressed as 50X, 100X. Coverage refers to the percentage of the target region actually read through sequencing, and is expressed as 98.3% and 99.4%.
In general, depending on where the inspection report is provided, it may be located at the front of the report or it may be located at the back of the report.
Methods
This section details the methodology and devices used for testing. We include the source of the sample, the sequencing devices used, and variant prioritization and interpretation methods to name a few.
Limitations
This section lists possible technical limitations of the test described in the Methods section. Depending on the capture kit, WES testing usually has limitations in not being able to read the entire exon regions, and variant detection may not be possible depending on the location and type of variant.
If you are curious about these limitations in detail, please refer to our blog post on the limitations of whole exome sequencing.
As described above, most laboratories that provide clinical genetic testing services follow ACMG guidelines, so the reports are likely very similar to our test report analyzed here.
Hopefully this will help you interpret clinical genetic test reports and more easily identify the essential information needed for your research or treatment of your patients, or use it as reference when delivering the test results to patients and their parents.
As genetic testing is becoming increasingly accessible and several genetic testing service providers are improving their reports to make them easier to interpret, we hope that through you, more patients with rare diseases can end their diagnostic odyssey.
ACMG Standards and Guidelines
The majority of clinical laboratories follow ACMG’s guidelines for interpreting genetic variants. To learn more about the guidelines, see What are the ACMG Standards and Guidelines and how do they work?
HGVS nomenclature
Variant names are written according to the nomenclature of the Human Genome Variation Society (HGVS). Learn how to read and write variants at the gene level and protein level on HGVS’s website.
Reference
- Skinner D, Raspberry KA, King M. The nuanced negative: Meanings of a negative diagnostic result in clinical exome sequencing. Sociol Health Illn. ;38(8):1303-1317. (2016).

What to Do After a Negative Genetic Test Result?
A negative result doesn’t always mean there’s no genetic cause. It might mean the answer is still hidden.
Get exclusive rare disease updates
from 3billion.

Yeonho Kang
Passionate about innovation and problem-solving, leading global growth marketing at a genetic testing provider, making a meaningful impact in rare disease care.







