Limitation of whole genome sequencing
Until now, we have mainly talked about the Whole Exome Sequencing (WES) -based diagnostic tests. However, as introduced in the ‘Limitation of Whole Exome Sequencing’, there are several limitations that exist with WES. Some exon regions are not covered well during target enrichment and PCR amplification processes of sequencing, and structural variants (SV) analysis with WES is limited. Whole Genome Sequencing (WGS) -based diagnostic tests can be a good alternative to overcome such shortcomings of WES. The result from meta-analysis study of chromosomal microarray analysis (CMA), WES and WGS as a first-line genomic test in children with intellectual and developmental disorder showed that WGS has the highest diagnostic utility.
Based on such evidence, many countries are striving to make rapid WGS (rWGS) a standard sick infant screening test. In the United States, 5 states have already made rWGS a Medicaid covered benefit.
However, is WGS, which covers all regions of the genome including the non-coding intron region really the perfect approach to genetic testing?
WGS is surely a superior approach in terms of being able to analyze SV, Copy Number Variants (CNV) and Mitochondrial variants that WES cannot properly analyze, but there are certain limitations to WGS that need to be addressed as well.
This article will discuss the limitations of WGS testing and the efforts to overcome these shortcomings.
Limitations
Higher cost of tests compared to WES
The cost of a WGS test is 2-3 times higher than the cost of WES. The amount of data produced from WGS is much larger to begin with. Hence, significantly more resources are required for data storage and analysis, which inevitably lead to cost differences. A review paper published in 2018 analyzed and compared the cost of WES and WGS from 36 research papers, and the result showed that despite the large difference between the minimum and maximum cost of both WES and WGS, when only the maximum costs of the two approaches were compared, the cost of WGS was 5 times higher than that of WES.
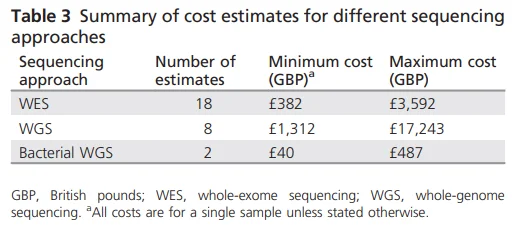
Figure 1. Summary of Cost estimates for different sequencing approaches
The amount of data produced for analysis in WES is approximately 10 Gb (150x) per patient, whereas the amount of data produced in WGS is 120 Gb (35x), which is 12 times larger. The numbers of variants that require interpretation differ by 60 times with 50 thousand variants for WES and 3 million for WGS. The average analysis time alone differs by 12 times with 2 hours required for WES and 24 hours for WGS. As such, WGS analysis requires more storage space, computing power and analysis time compared to WES, resulting in large cost differences.
Challenges of variant interpreting due to lack of information
85% of the disease causing genetic variants are known to be located in the exon region, and while there have been many studies conducted on the exon region, our understanding of the non-coding region is still lacking. Additional variants discovered by WGS include many non-coding region variants with not enough evidences for variant classification. Many non-coding variant interpretations must rely on in silico prediction alone. It causes confusions among clinical geneticists in identifying the pathogenic variants. In other words, there is the limitation that even with the complete genome sequence, there is still insufficient evidence to classify or interpret the detected variants. Other types of diagnostic tests may be much more effective in patients with clear symptoms that are associated with a specific disease.
Fundamental limitation of the short-read sequencing technology
Unlike WES, WGS does not include PCR amplification and target enrichment processes. This gives WGS a great advantage in that it can read the entire sequence with uniform depth and prevent bias from variations in coverage. However, both approaches use the short-read sequencing technology of fragmenting the DNA to 300bp~400bp reads for sequencing, and has strong dependency on the reference genome. Due to such nature of the technology, there is a limitation in analyzing structural variants of 300~400bp or longer repeats. Also, there is limitation to analyzing large deletion and insertion.
3 approaches to reducing the cost of WGS testing
Selecting patients who are most likely to benefit from WGS
WGS tests are recommended when it is difficult to narrow down to specific rare diseases due to a wide range of symptoms, and when no pathogenic variants have been found from other diagnostic approaches such as NGS panel tests or WES tests etc.
rWGS for newborns suspected of rare disease
rWGS is already covered by Medicaid in 5 states for newborns suspected of rare diseases. The diagnostic and clinical utility of rWGS has long been established by multiple researches, and last January its cost effectiveness was also confirmed through a research.
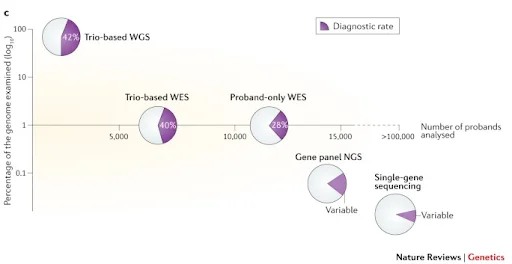
Figure 2. Cost Efficacy of Rapid Whole Genome Sequencing in the Pediatric Intensive Care Unit
Reduced cost of WGS test improves its accessibility
Cost of WGS test includes costs required for sequencing, data storage, data analysis etc. Owing to technological advances, all of these costs are continually dropping.
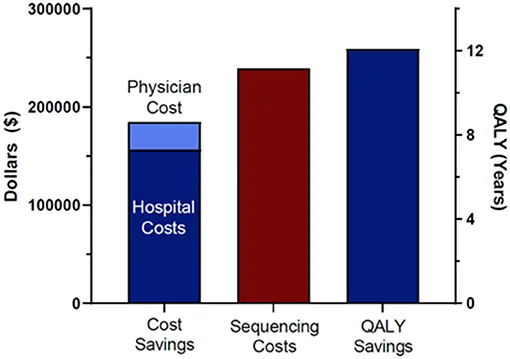
Figure 3. Advantages and Perils of Clinical Whole-Exome and Whole-Genome Sequencing in Cardiomyopathy
The above graph demonstrates the costs of targeted gene panel test, a cheaper NGS test analyzing only a set of selected genes, WES and WGS (Figure 3). NGS tests (WES, WGS) that previously had low accessibility due to high cost have become more affordable, and it is expected to replace existing panel tests in the near future by matching their cost level. Companies should continue their efforts to lower the cost and increase the accessibility of WGS by improving predictive performance with accumulated data, and reducing resources for data analysis through automatization.
Moreover, insurance coverage for WGS tests is expected to be expanded. 12 insurance companies, covering around 162 million people in United States, responded to an interview that they may review expansion of coverage for WGS once more evidences supporting its clinical utility are collected and the way they help with coverage decision making is improved.
3 approaches to interpret difficult information.
Growing pace of research in disease-associated genes
Owing to the advancements of science and technology, study on gene-phenotype relationships are rapidly accumulating and being shared through online. The figure below, which was also introduced in the previous blog posting, shows the increasing trend of OMIM database that is widely used in rare disease diagnostic. As of June 8th, 2022, total number of phenotypes for which the molecular basis is known was 7,188 and total number of genes with phenotype-causing mutation was 4,648. This information has great impact on the diagnosis of rare diseases.
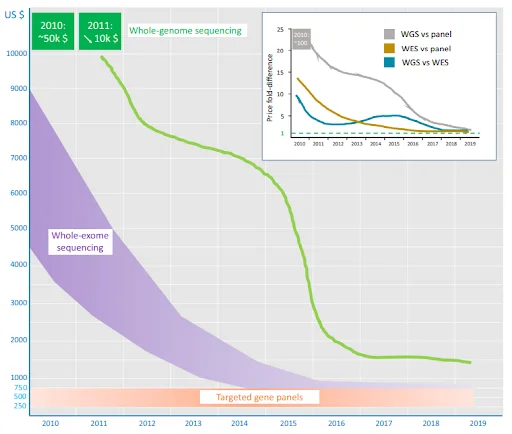
Figure 4. OMIM.org: leveraging knowledge across phenotype–gene relationships
Predicting pathogenicity of gene variants using AI Technology
Average of 3 million variants are detected from WGS. It is physically impossible to collect solid evidence on the effect of each or a combination of these genetic variants. For this reason, there are researches underway to use algorithms and artificial intelligence technology in predicting pathogenicity of such variants. However, there are very diverse types of rare diseases present and the number of patients for each disease is limited, which makes it challenging to collect enough data to train an AI. 3Cnet, an AI model developed by 3billion, showed high performance in predicting the pathogenicity of variants by utilizing clinical data from ClinVar, common variant information from GnomAD and conservation data from UniRet.
3billion also has a history of being awarded as the top performing team in 6th Critical Assessment of Genome Interpretation (CAGI6), a global artificial intelligence genomic interpretation contest.
Precise variant screening through Trio-based tests
When requesting for WGS, using a trio-based test that screens not only the proband but also both parents can be more effective in detecting de novo variant and pathogenic variants. Selecting trio-based WGS can be a good way to reduce the ultimate cost required to obtain a diagnosis.
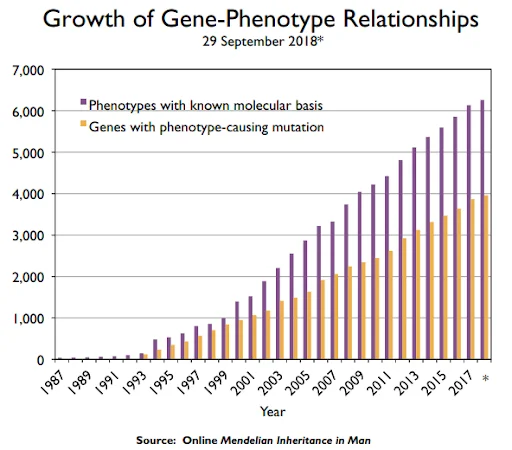
Figure 5. Paediatric genomics: diagnosing rare disease in children
The above research outcome shows that trio-based tests improve the diagnostic yield in both WES and WGS.
2 approaches to overcome technological limitations.
Advancement of Long-Read Sequencing Technology
Long-Read Sequencing(LRS) method that can overcome the technological limitations of Short-Read Sequencing(SRS) is under development. SMRT by PacBio and nanopore sequencing by ONT are some of the representative examples. As can be seen by the below figure, long-read sequencing technology has advanced rapidly over the past decade.
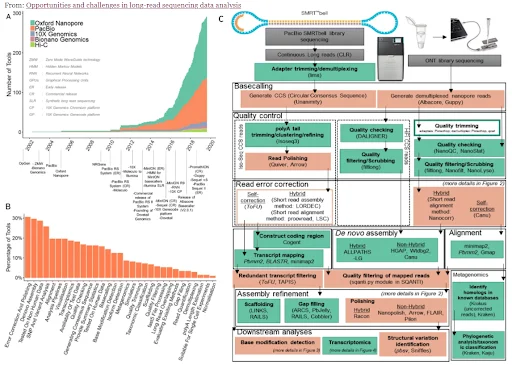
Figure 6. Opportunities and challenges in long-read sequencing data analysis
LRS can read DNA fragments of ≥1,000bp up to 1 million base pair, and provide information on DNA methylation. In comparison to SRS, LRS can improve de novo assembly, mapping certainty, transcript isoform identification and structural variant analysis.
In a paper published on AJHG in 2021, LRS analysis was performed on Oxford Nanopore platform on 40 patients. Among the 40 patients, 10 individuals failed to find pathogenic variants despite already going through multiple diagnostic tests. In 6 of these patients pathogenic or likely pathogenic variants were detected and in 2 patients variants of uncertain significance were detected. With the help of LRS, more detailed information was able to be obtained about the variants that were previously detected from other diagnostic approaches, and pathogenic variants that were previously undetected from other tests were discovered.
However, high error rate, low accuracy and high cost are some of the limitations of LRS that still remain to be solved.
Combination of Transcriptome and Genome testing
Even if there is a pathogenic variant present in genome, only when transcription occurs in that region, does the genetic variant lead to the expression of phenotype and cause the disease. Therefore, by conducting transcriptome analysis, which has a more direct effect on phenotype expression, in parallel with WGS can increase the diagnostic yield. According to the paper published by Dr. Hane Lee, current CGO (Chief Genomic Officer) of 3billion, in 2020 during her tenure at UCLA as a professor, performing WES/WGS together with RNAseq had a diagnostic yield of 38%. And in 18% of the total cases of diagnosis, RNAseq played an essential role in determining the pathogenicity of the variant.
As such, rather than performing genetic tests alone, conducting additional tests that can track post-transcriptional processes in combination can help improve the diagnostic rate.
Conclusion
Up to now, 3billion has been focusing on a WES-based diagnostic test(3B-EXOME). But starting on June 14th, we have launched a WGS-based diagnostic test(3B-GENOME) for rare disease patients with hopes of delivering successful diagnosis to more patients.
As a diagnostics company, it is important that we introduce the limitations and problems of WGS as well as promoting WGS and its advantages. Even with the world-class diagnostic capability, there are still a lot of challenges to be overcome to complete a perfect diagnostic tool. 3billion will continue to strive to increase accessibility to diagnostics and improve diagnostic rates until rare disease patients cease to suffer from diagnostic odyssey.

Discover Our Advanced Whole Genome Sequencing
Get the most detailed and accurate Genome Test available.
Get exclusive rare disease updates
from 3billion.

Taeyeon Bae
Marketer & Growth hacker at 3billion. We are using a data-driven approach to improve the lives of people with rare diseases.