Getting started with the rare disease database: OMIM

Are you curious about which rare disease is related to a particular gene? Or, want to find out the phenotype, inheritance pattern, and historical information of a specific rare disease? OMIM database is for you!
OMIM database (https://omim.org, shortened for Online Mendelian Inheritance in Man) is a collection of data about human gene-phenotype relationships, genetic disorders, and their characteristics. Based on literature curated by experts, it provides information on over 7000 genetic disorders and its associated genes, symptoms, inheritance pattern, age of onset, etc. And the database is updated every day for new gene-phenotype relationships.
First of all, before you use the OMIM database, you need to know the numbering system specified in OMIM. Every gene or disease/phenotype is called an ‘entry’ and each entry has a unique 6 digit number ID called the MIM number. The meaning of the first number of the 6 digits of a MIM number is as follows1:
- 1—– (100000- ) 2—– (200000- ): Autosomal loci or phenotypes (entries created before May 15, 1994)
- 3—– (300000- ): X-linked loci or phenotypes
- 4—– (400000- ): Y-linked loci or phenotypes
- 5—– (500000- ): Mitochondrial loci or phenotypes
- 6—– (600000- ): Autosomal loci or phenotypes (entries created after May 15, 1994)
Also, each MIM ID has a symbol. Symbols refer to the character attached to the front of the MIM ID. The types of symbol are as follows1:
- An asterisk (*) before an entry number indicates a gene.
- A number symbol (#) before an entry number indicates that it is a descriptive entry, usually of a phenotype, and does not represent a unique locus. The reason for the use of the number symbol is given in the first paragraph of the entry. Discussion of any gene(s) related to the phenotype resides in another entry(ies) as described in the first paragraph.
- A plus sign (+) before an entry number indicates that the entry contains the description of a gene of known sequence and a phenotype.
- A percent sign (%) before an entry number indicates that the entry describes a confirmed mendelian phenotype or phenotypic locus for which the underlying molecular basis is not known.
- No symbol before an entry number generally indicates a description of a phenotype for which the mendelian basis, although suspected, has not been clearly established or that the separateness of this phenotype from that in another entry is unclear.
- A caret (^) before an entry number means the entry no longer exists because it was removed from the database or moved to another entry as indicated.
Let’s take a closer look at the database!
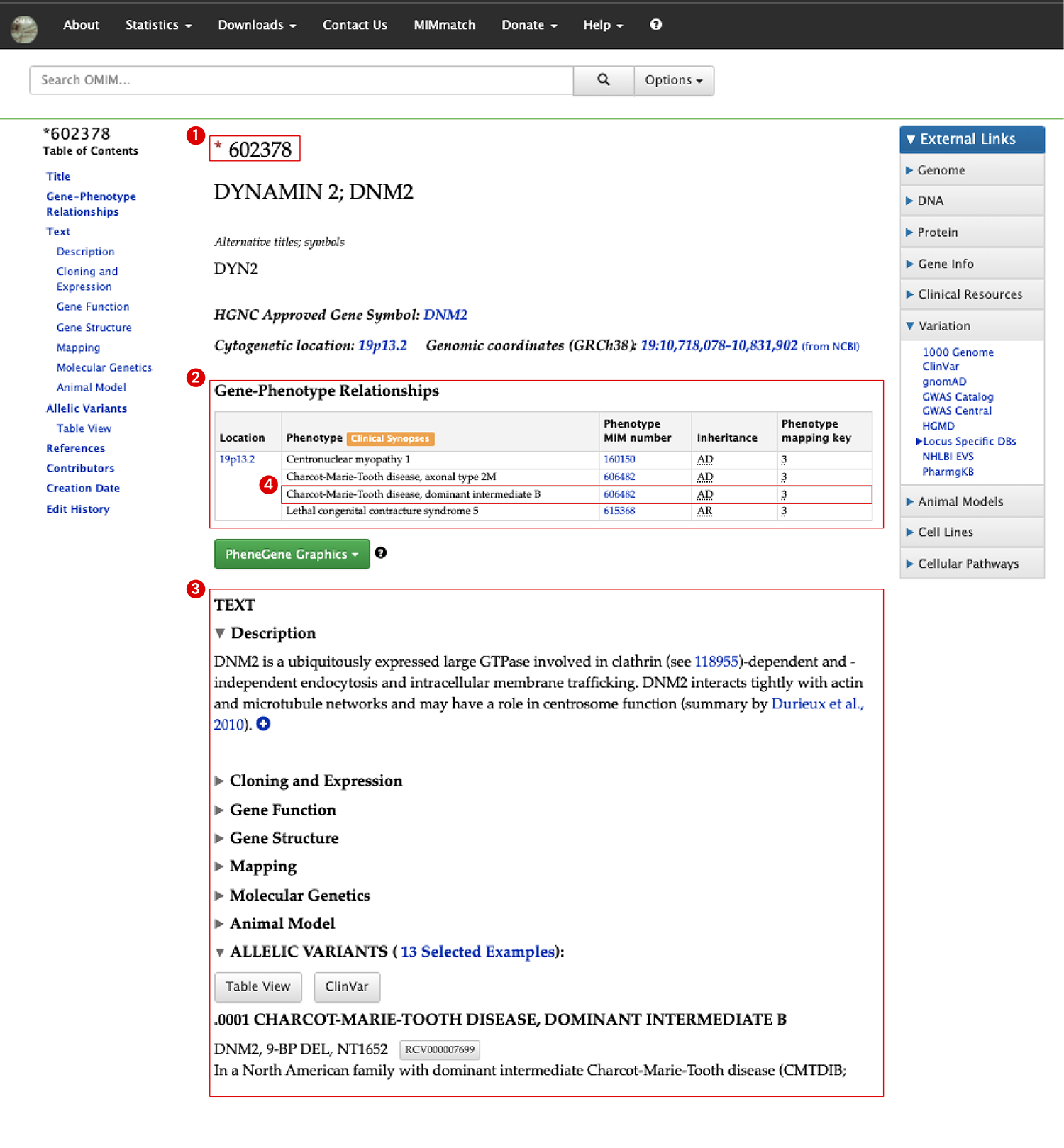
This is a screenshot of the page that appears when you search DNM2 on the OMIM website. The page can be divided into 3 main sections: ‘Title’, ‘Gene-Phenotype Relationships’, and ‘Text’ sections. As you can see on the top of the page in the ‘Title’ section, it has a MIM number of 602378, and since an asterisk(*) symbol is on its left side, this ID indicates a ‘gene’ entry (See 1 in Image 1). Under the title and alternative names, there’s a ‘Gene-Phenotype Relationships’ section in which phenotypes/disorders that are related to the DNM2 gene are listed with its inheritance (See 2 in Image 1). At the bottom, in the ‘Text’ section, a description of the gene, the experimental functions of the gene reported in papers, and links to the associated variants (‘Allelic Variants’) reported in ClinVar are provided (See 3 in Image 1).
Let’s click on one of the phenotypes, 606482: Charcot-Marie-Tooth disease as an example here, in the Gene-Phenotype Relationships section (See 4 in Image 1).
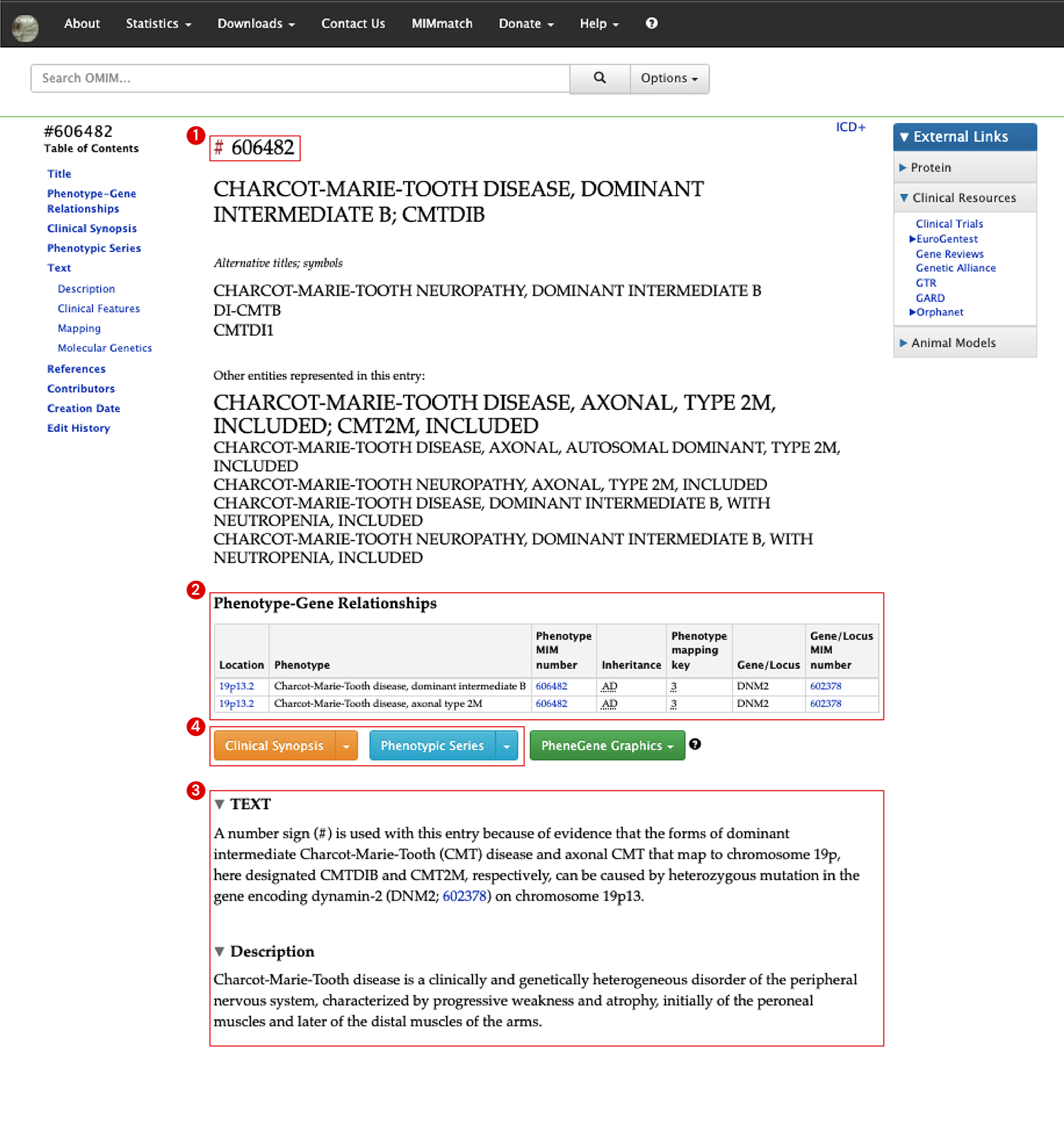
Likewise, the unique MIM ID of Charcot-Marie-Tooth disease, 606482, is at the top and the number symbol (#) indicates this entry is a phenotype (See 1 in Image 2). In the ‘Phenotype-Gene Relationships’ table, you can also see the link to DNM2 gene and inheritance pattern (See 2 in Image 2). A description of clinical features and relationships can be found in the ‘Text’ section (See 3 in Image 2).
Additionally, on the phenotype page, there are separate sections for ‘Clinical Synopsis’ and ‘Phenotypic Series’ which are not found on the previous page (gene page) (See 4 in Image 2).
So let’s take a look at each section.
First, If you open the ‘Clinical Synopsis’ section, you can see the symptoms of this disease as shown in the screenshot below.
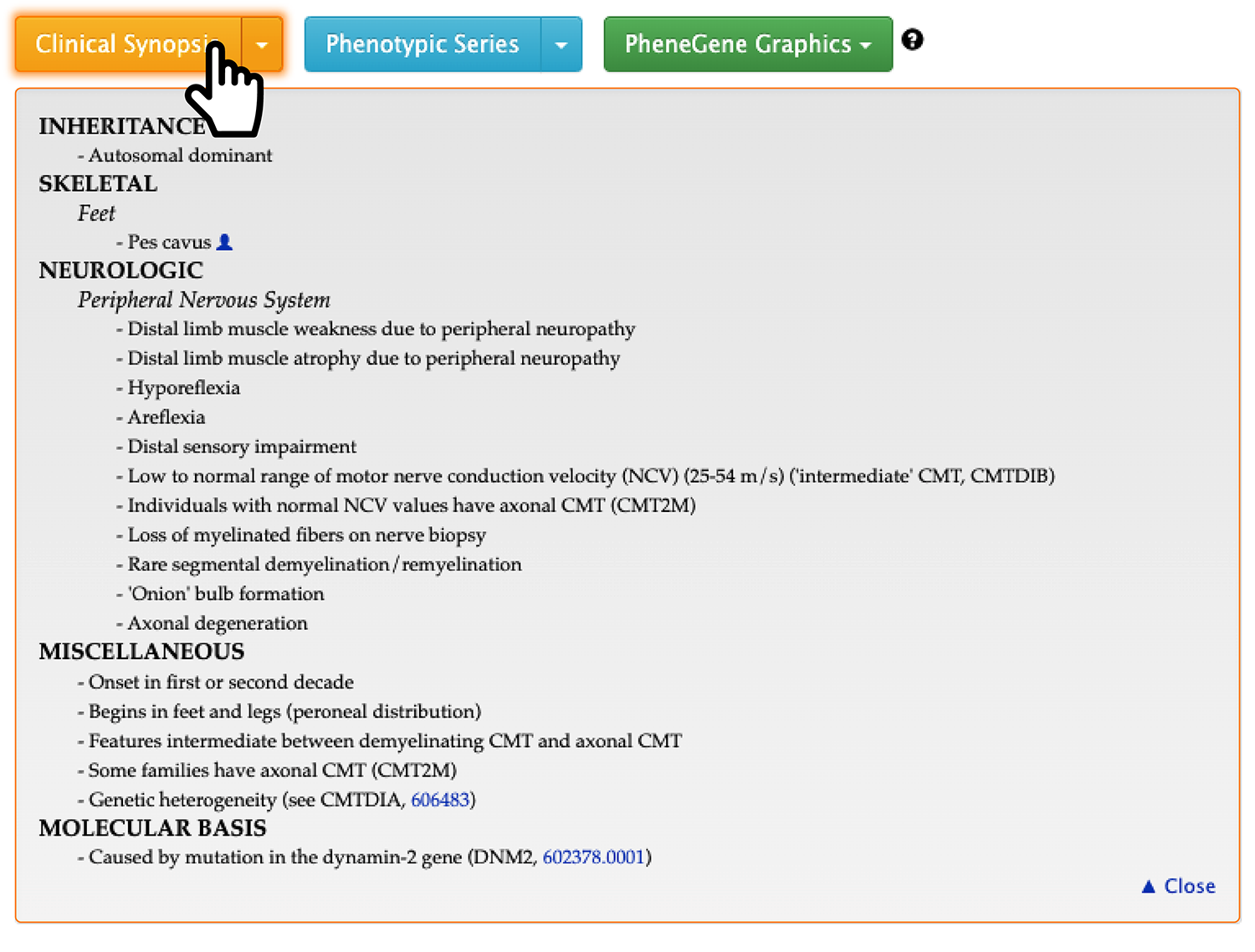
Furthermore, if you hover your mouse over the icon of a person, you can also see the actual morphology picture reported in the paper.
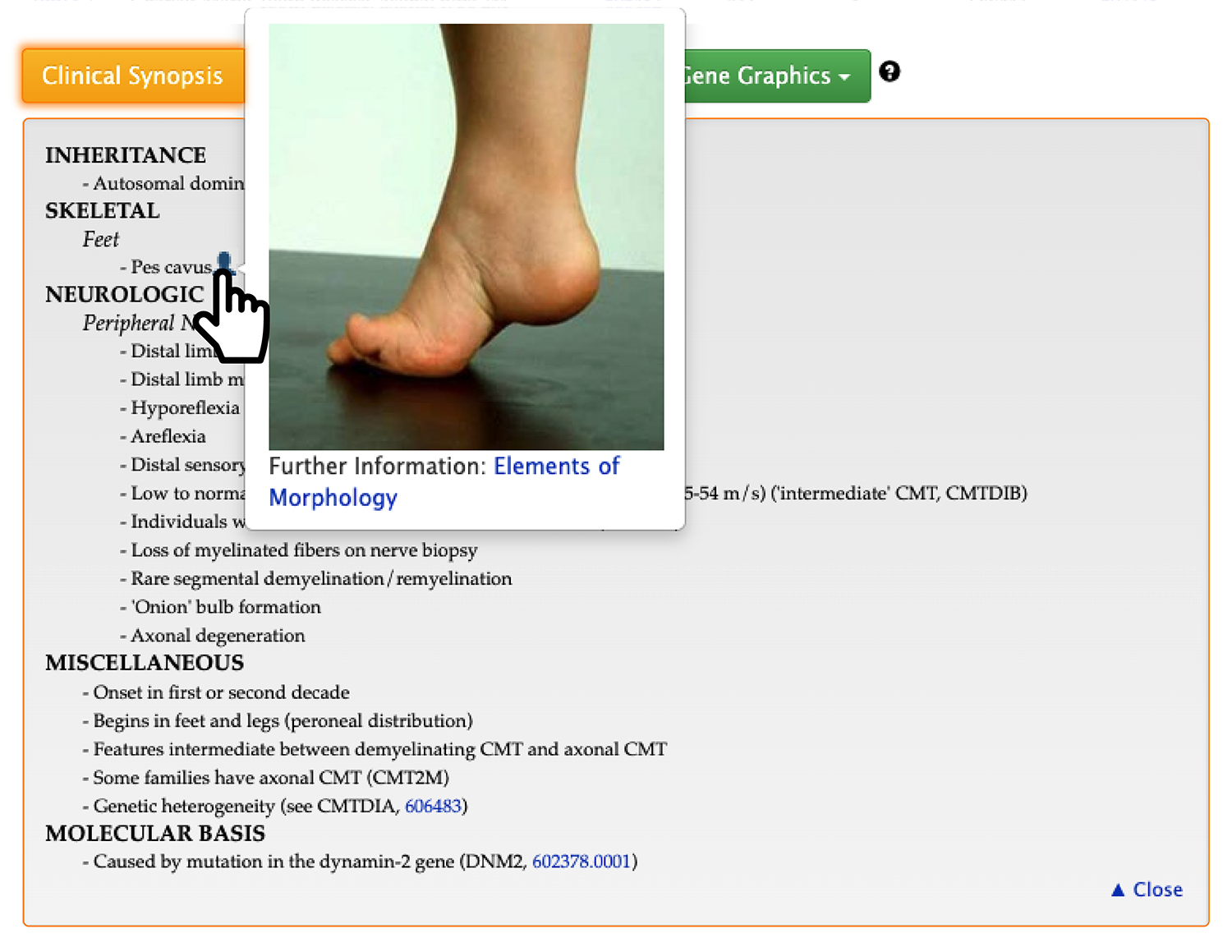
Second, if you expand the ‘Phenotypic Series’ section, you will see a table like the following.
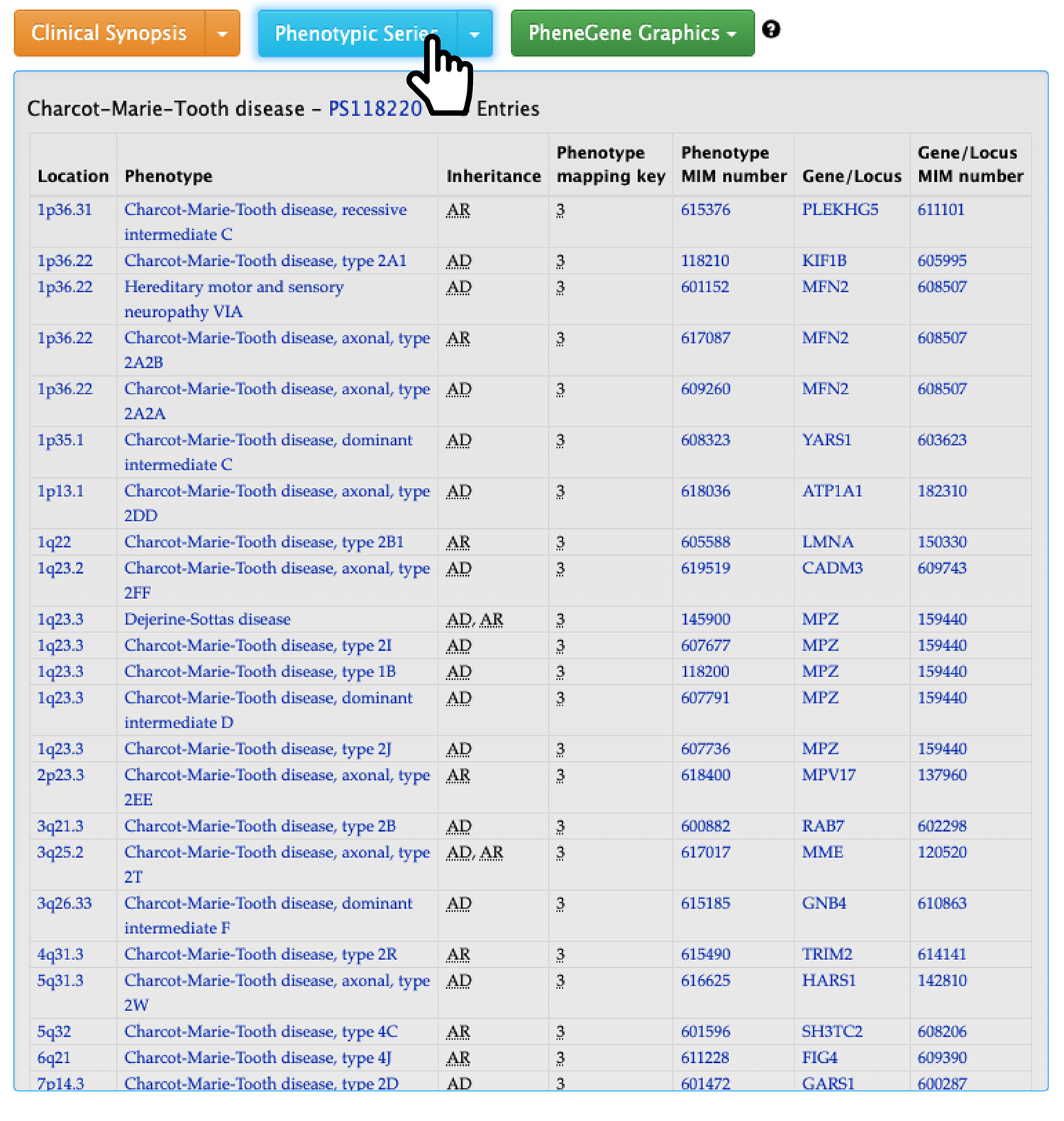
Phenotypic series is the genetic heterogeneity of similar phenotypes across the genome. It has the prefix of ‘PS’ followed by 6 numbers. Therefore, a list of all phenotypes and genes bound to PS118220 is shown.
We briefly looked at each section of the OMIM database. Now you will be able to find information about the gene or disease that you’re interested in. However, there are many gene-diseases that have not yet been identified and unknown diseases that are not included. So it is important to be aware and not judge the diseases in OMIM as absolute.
As a user, I think the one thing this database lacks is that the symptoms listed in the ‘Clinical Synopsis’ section or in the description section are not mapped to the HPO term. HPO (Human Phenotype Ontology)2 term is the standardized term of phenotypic abnormalities in human disease and is universally used in diagnostics (3billion uses HPO terms for symptoms!). Although there are external links to ClinVar, UniProt, or other databases (at the top right corner of the page) provided as recent features, I believe it would be more helpful to link the symptoms to the HPO database. Hopefully this will be added in an update in the near future 🙂
References
- Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). World Wide Web URL: https://omim.org/
- Sebastian Köhler., et al., The Human Phenotype Ontology in 2021. Nucleic Acids Research, Volume 49, Issue D1, 8 January 2021, Pages D1207–D1217, https://doi.org/10.1093/nar/gkaa1043

Curious how genetic data leads to real diagnoses?
Explore how we turn genomic insights into clinical action. Get the full overview of our diagnostic services—including exome, genome, and data interpretation.
Get exclusive rare disease updates
from 3billion.

You Kyung Cho
Bioinformatics expert and full-stack developer at 3billion, creating web solutions that enhance rare disease diagnostics, bridging data science with UX design.







