Gene Coverage in WES: The Hidden Gaps Clinicians Should Know
When choosing a genetic testing method, many clinicians and researchers turn to Whole Exome Sequencing (WES) for its broad coverage and cost-efficiency. In a previous article, we explored the differences between targeted panels and WES, concluding that WES can (almost) encompass the scope of most NGS panels. But here comes the natural follow-up question:
Does WES reliably cover every gene without issue?
Unfortunately, the answer is no—and the reason lies in the technological limitations of next-generation sequencing (NGS) platforms.
The Limits of Short-Read Sequencing
Most clinical-grade NGS today is powered by short-read sequencing technologies, with Illumina being the most commonly used platform. These systems read DNA in short fragments, typically around 150 base pairs (bp) in length.
While this is sufficient for a majority of the genome, certain regions pose significant challenges:
While this is sufficient for a majority of the genome, certain regions pose significant challenges:
- Homologous genes: Genes that share nearly identical sequences, like SMN1 and SMN2, are difficult to distinguish in short-read data. Misalignment or ambiguous mapping can lead to unreliable variant calls.
- Repeat expansions: Tracts of repetitive DNA—such as those implicated in Huntington’s disease or fragile X syndrome—are notoriously difficult to resolve using short reads.
- GC-rich regions: High GC content can hinder the efficiency of PCR amplification and sequencing, leading to dropouts or low coverage.
Why It Matters
In clinical diagnostics, missing a variant due to insufficient coverage can have serious consequences. For genes known to play a role in disease, ensuring reliable coverage is essential—especially in rare disease diagnosis where every variant counts.
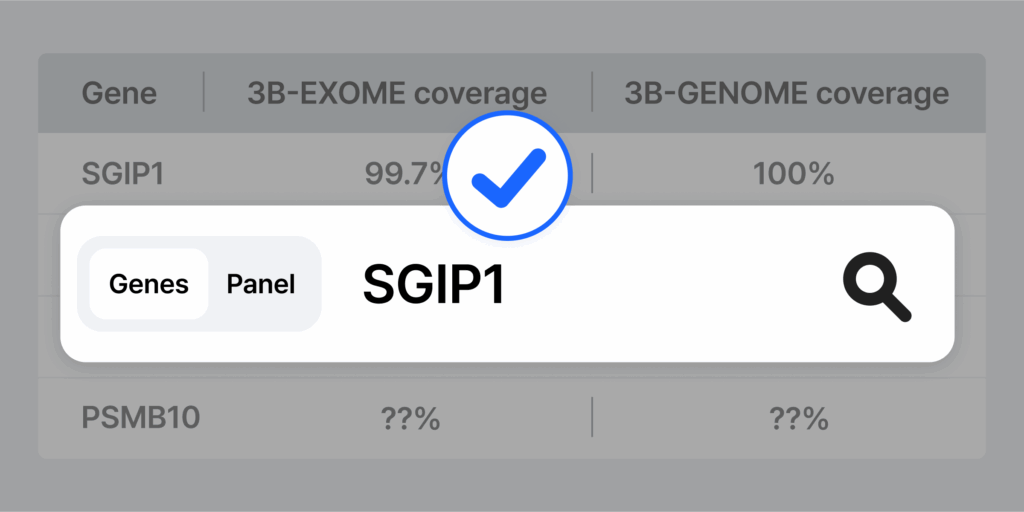
How to Check Gene-Level Coverage
Wondering whether a specific gene is well-covered in WES or WGS? You don’t have to guess.
At 3billion, we provide a free tool called the Gene Coverage Browser, where users can explore gene-level coverage data from real sequencing runs. It’s designed to help clinicians, researchers, and lab directors make informed decisions about test selection and limitations.
Get exclusive rare disease updates
from 3billion.

Sree Ramya Gunukula
Marketing Leader with experience in the pharma and healthcare sectors, specializing in digital health, genetic testing, and rare disease diagnostics.







