How 3billion’s WES Overcomes Exome Sequencing Limitations
Exome sequencing (ES) has established itself as a pivotal tool in molecular diagnostics due to its cost-effectiveness and ability to identify pathogenic variants within coding exons, where approximately 80% of reported pathogenic variants reside. Despite its advantages, ES faces several limitations.
Limitations of Standard Exome Sequencing
- Coverage Uniformity: Unlike genome sequencing(GS), exome sequencing can suffer from uneven coverage, especially in GC-rich regions(where many C and G building blocks are present) or repetitive regions(where the same pattern repeats many times) due to Polymerase Chain Reaction bias(PCR Bias). This can result in missed pathogenic variants in these critical areas.
- Non-coding Regions: ES focuses on coding regions but ignores the parts of the non-coding regions. However, sometimes, these ignored parts can also have changes that cause diseases, so missing them can limit the diagnostic yield.
- Copy Number Variant (CNV) Resolution: Exome sequencing has reduced sensitivity in detecting copy number variants compared to genome sequencing, making it challenging to identify complex structural variants. CNVs can involve large sections of the genome, and the resolution of ES is typically insufficient for reliable detection.
- MT Genome Coverage: ES generally offers low coverage of the mitochondrial genome unless specifically targeted, necessitating separate tests for comprehensive analysis.
For a more detailed exploration of the limitations of exome sequencing, please refer to the comprehensive article on the Limitations of WES.
Despite these limitations, ES remains more affordable than GS. The challenge is to mitigate these drawbacks without incurring the higher costs associated with GS.
Customization Approach for Enhanced Exome Sequencing Coverage
“Exome Boosting by 3billion” addresses these limitations through a customized exome capture probe design to augment coverage in several key areas.
- Coding Exons with Insufficient Coverage: By addressing the insufficiencies in exon coverage, 3billion’s Enhanced Exome significantly enhances the reliability and comprehensiveness of genetic diagnostics, ensuring that critical pathogenic variants are noticed. For instance, the ORF15 exon of the RPGR gene is a well-known drop-out region in standard exome sequencing, often resulting in insufficient coverage and a higher risk of missing pathogenic variants. The RPGR gene is associated with several severe ocular conditions, including retinitis pigmentosa, cone-rod dystrophy, and macular degeneration. In the original 3B-EXOME, the coverage could be more sparse and consistent. In contrast, the Enhanced 3B-EXOME provides comprehensive and uniform coverage across the entire ORF15 exon, ensuring reliable detection of any pathogenic variants present.
- Non-coding Regions: In the enhanced version, additional capture baits have been added across non-coding regions that harbor disease-causing variants. This results in significantly improved coverage and detection capabilities for these non-coding regions, as illustrated by the more comprehensive and consistent read depth in the enhanced 3B-EXOME compared to the original. Regular updates to the capture bait design ensure newly identified non-coding variants are also targeted, providing a more comprehensive diagnostic tool.
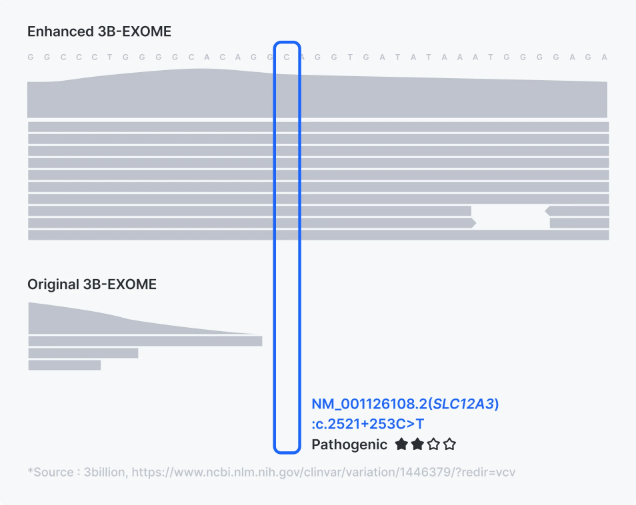 Improved coverage and detection capabilities for the non-coding regions in 3billion enhanced exome sequencing.
Improved coverage and detection capabilities for the non-coding regions in 3billion enhanced exome sequencing. - Mitochondrial Genome: In the enhanced version of 3billion’s exome testing, the coverage of the mitochondrial genome has been significantly increased. This integration within the exome sequencing process eliminates the need for separate mtDNA tests, which can be time-consuming and costly and ensures that mitochondrial variants are detected alongside nuclear DNA variants without additional procedures. For further details, please refer to the comprehensive article on Enhanced WES.
Evidence of Efficacy in Exome Sequencing Improvements
The impact of Exome Boosting is evident in the clinical results observed over three months.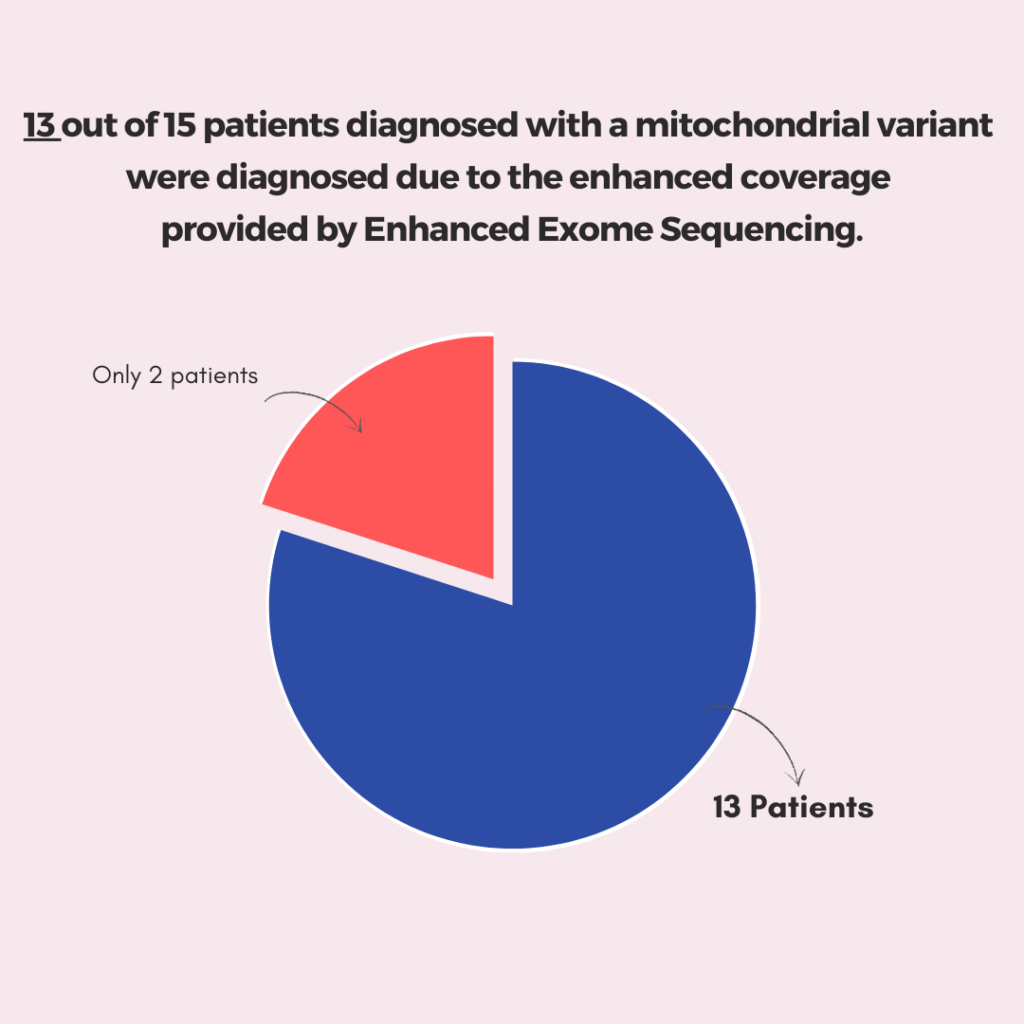 13 out of 15 patients diagnosed through enhanced coverage in 3billion’s Whole Exome Sequencing
13 out of 15 patients diagnosed through enhanced coverage in 3billion’s Whole Exome Sequencing
- Out of 1,388 patients with diagnostic findings, 26 patients (1.9%) were diagnosed due to the enhanced coverage provided by Exome Boosting.
- 13 out of 15 patients diagnosed with a mitochondrial variant were found to have heteroplasmic variants, demonstrating the efficacy of the augmented mitochondrial genome coverage.
- 13 out of 1,373 patients were diagnosed with 14 single nucleotide variants (SNVs) and insertions/deletions (INDELs) within the boosted regions, including:
- A stop-gained variant in FRMPD4.
- Variants in the RPGR ORF15 region are known for its many pathogenic variants and traditionally poor exome capture.
- Variants in SNORD118, a non-coding gene.
- Intronic variants in GHR and HBB.
- A 3’UTR variant in PHEX.
3billion’s Future Plans for Enhanced Exome Sequencing and Improved Genetic Diagnostics
3billion is committed to continuously updating the custom hybridization panel to:
- Normalize the coverage across the augmented regions, ensuring consistent and comprehensive analysis.
- Add newly discovered genes and non-coding regions that contain newly submitted or reclassified ClinVar pathogenic/likely pathogenic (P/LP) variants.
In conclusion, this enhanced exome sequencing from 3billion significantly improves the reliability and comprehensiveness of genetic diagnostics, ensuring critical pathogenic variants are detected. Experience the future of genetic testing with 3billion’s Enhanced Exome Sequencing and ensure no pathogenic variant goes undetected—contact us today to learn more.
Get exclusive rare disease updates
from 3billion.

Sookjin Lee
Expert in integrating cutting-edge genomic healthcare technologies with market needs. With 15+ years of experience, driving impactful changes in global healthcare.







