Simple Guide to Privacy-Preserving Techniques for Sharing Genomic Data
Summary
- The increasing integration of multiple datasets for a wide range of studies also poses significant privacy concerns.
- Even if data are processed under privacy regulations such as HIPPA, data privacy cannot be perfectly guaranteed.
- Privacy preserving techniques including data anonymization such as k-anonymity, or differential privacy (DP) can be used to reduce privacy risk in shared data.
- Privacy preserving techniques can reduce the risk from unexpected privacy infringement after releasing public data
Keywords: genomic data privacy, anonymization, deidentification, differential privacy
Introduction: the importance of genomic data privacy
Several research initiatives are combining genomic data with other types of data, such as clinical data. In surveys, many researchers need to integrate various data into their research field for new scientific findings1. To integrate various data including genomic data, data sharing is required by collecting data vertically (collecting same-formatted data) or horizontally (collecting various data modalities). However, it raises privacy concerns in the process.
Owing to the characteristics of biomedical data, such as immutability, processing genomic data requires extra caution. For example, unlike identification numbers such as social security numbers (SSN), genomic data is immutable. As a result, the leakage of genomic data constitutes a high risk of breach of privacy. To address this issue, removing identifiers was used to share data with the public conventionally. Unfortunately, sanitization does not ensure deidentification.
An individual could be identified even after identifiers were removed in one case. In Massachusetts, a government agency called GIC (Group Insurance Commission) that provides health insurance to state employees released hospital visits for research purposes. The fields containing name, address, SSN, and other explicit identifiers, were removed by the government agency. GIC assumed it had protected employees’ privacy; however, a graduate student identified a governor’s records by combining the prior knowledge that the governor lived in Cambridge with voter rolls containing name, ZIP codes, birth dates, and gender2. This is an example of a linking attack, in which auxiliary data is combined to reidentify an individual3.
To reduce privacy infringement as possible, several countries have implemented regulations to protect people’s privacy. For example, in order to share data, healthcare organization must follow their privacy rule or act, such as the Health Insurance Portability and Accountability Act (HIPPA) or Genetic Information Nondiscrimination Act (GINA)4. However, compliance with these regulations which require appropriate safeguard to protect the privacy does not completely guarantee data security. The possibility of privacy invasion still exists. This is because individuals could be identified with the advanced data processing technique.
Once the privacy of genomic data is infringed, the privacy exposure is permanent because genomic data cannot be changed like other biomedical information. Therefore, organizations or agencies that handle genomic data must follow privacy rules and regulations. This article introduces privacy-preserving techniques. Understanding these can help to avoid a privacy leakage disaster and improve privacy policies or security techniques.
Basic concepts related to data privacy
Before delving into privacy techniques, it is important to understand the definitions of key terms.
- Privacy risk: It usually refers to the likelihood that individuals would be identified. The privacy risk is relative and is determined by the attacker’s auxiliary information. As an example, if the student is unable to consider external information, a governor may not have been identified if GIC released health records.
- Deidentification and Anonymization: Deidentification is the process of permanently erasing identifiable data. According to the International Standard Organization (ISO), deidentification refers to the process of deleting identifiers, which includes anonymization and pseudonymization. Especially anonymization refers to the process by which personally identifiable information is irreversibly altered or deleted such that it can no longer be identified. It emphasizes the permanent modification of identifiers. In terms of removing identifiers, both are similar; however, deidentification does not specify the permanent deletion of identifiers because it includes pseudonymization. Deidentification is commonly accomplished by deleting a person’s name or SSN5,6,7.
- Pseudonymization: It is the process of creating a fictitious name. The modification of identifiers is not permanent, but it is reversible. This is a method of assigning a fake identifier to an individual, removing the identifier from the original data, and retaining pairs of fake and real identifiers. As a result, a data processor with the option of privacy can reidentify. In 3billion, for example, in-house generated IDs (pseudonymized ID) are used to replace patient identifiers such as patient names. This pair can only be viewed by authorized users.
- Identifier, direct identifier, indirect identifier, and quasi-identifier: Direct and indirect identifiers are the two types of identifiers. Direct identifier is an attribute that can be used to uniquely identify a data subject on its own. In contrast, an indirect identifier is an attribute that allows a data subject to be identified with other attributes from external or given data. In particular, a quasi-identifier is an indirect identifier in a given dataset (dataset context)8. For example, a SSN is a direct identifier. In addition, suppose we have a medical history dataset with an address code and external claim dataset including age, gender, diagnosis code, and address code. Age, gender, and diagnosis code can all be used as indirect identifiers. Also, attributes which can identify individuals with other attributes in medical history dataset could be quasi-identifier (dataset context).
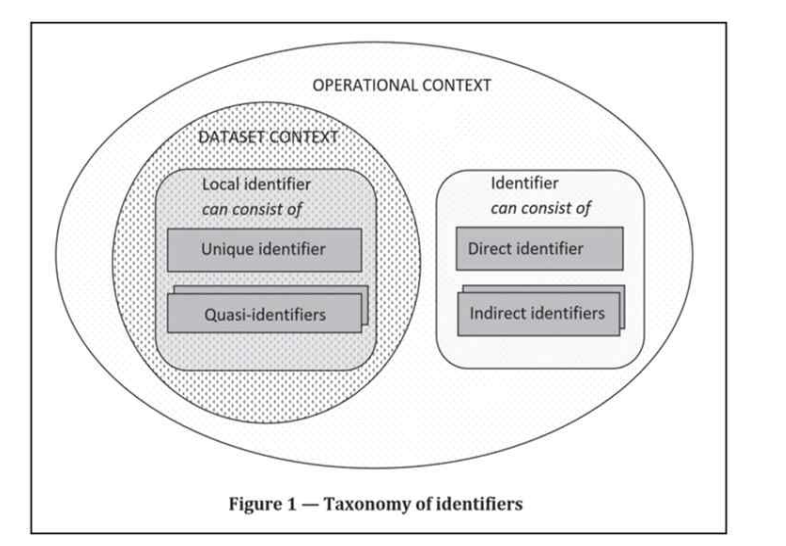
Figure 1. Taxonomy of identifiers
This article introduced the definitions of the basic concepts related to privacy; however, it may be difficult to classify which data fall into these categories. Regardless, understanding them can assist you in developing a deidentification strategy and data processing procedures.
Privacy-preserving technique
1. Data anonymization: irreversibly changing or deleting identifiers
- k-anonymization: k-anonymity is commonly used for data anonymization; k-anonymity is a property of data, not a technique. A dataset is said to have k-anonymity if there are no rows with the same attributes that are less than k. It is also known as the power of “hiding in the crowd.” Suppression and generalization can be used to achieve k-anonymity in practice. Suppression refers to the replacement of attributes with masking characters, whereas generalization refers to the replacement of attributes with boundary categories.
- As shown below, dataset D can contain multiple rows. If only two rows with ages of 20 exist in dataset D, the dataset does not have three levels of anonymity. To achieve 3-anonymity in the dataset, we can suppress the (right upper) columns by replacing age columns with “*.”
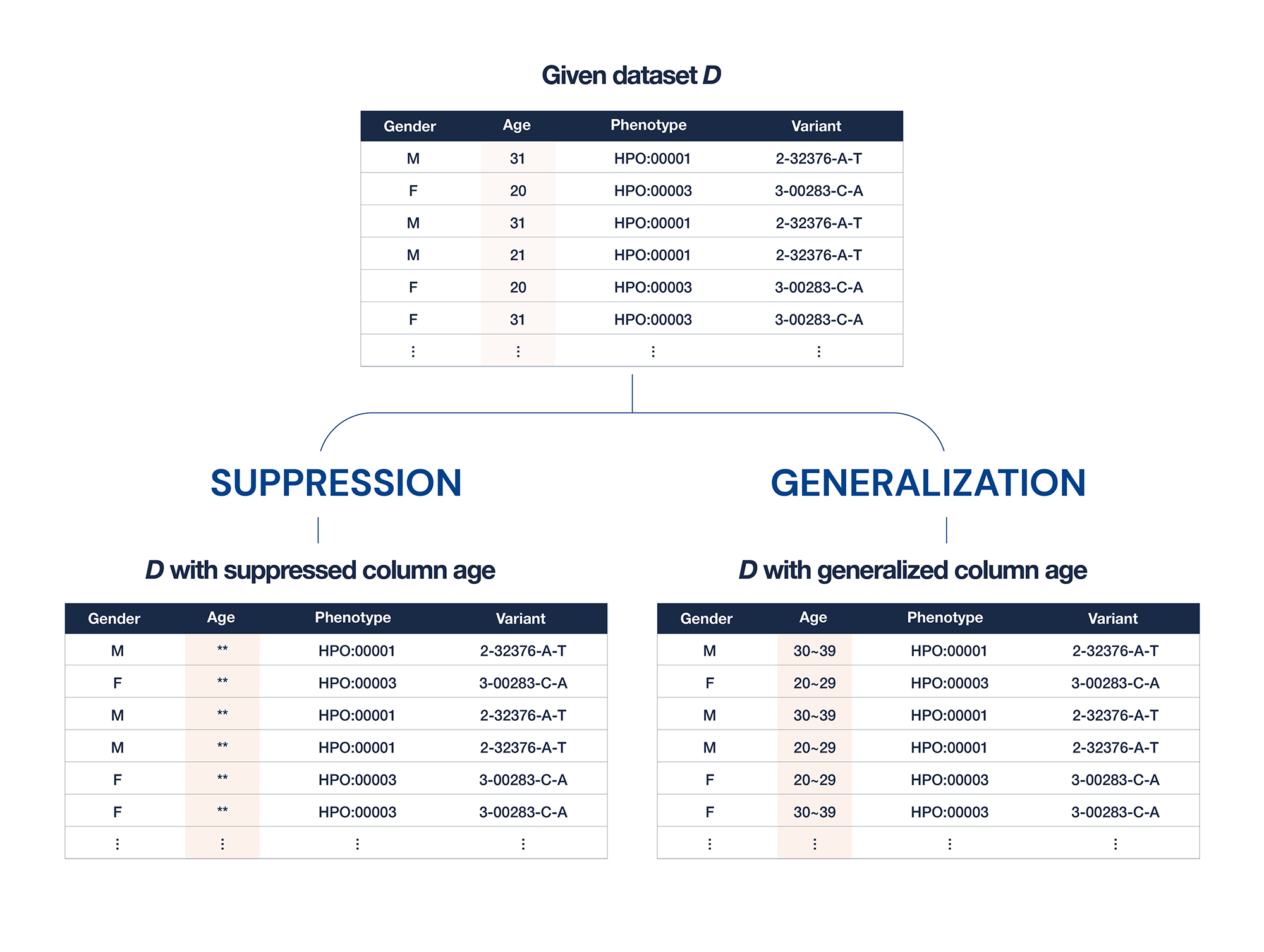
Figure 2. Illustration of two typical method for k-anonymization
In addition, as shown in Figure 2, we can modify the age columns with categorical variables.
2. Differential privacy (DP): DP causes statistics (adding noise) from a specific distribution to be perturbed. DP, in contrast to k-anonymity, is a property of algorithms. The term “differential privacy” refers to the difference in records between two datasets.
Assume that an institution creates a public database and makes it available to unspecified people. This database provides statistics on records, not records they want in the database. As a result, researchers can obtain the statistics of eligible participants rather than the entire dataset from database by entering conditions. We can attack the privacy of individuals in datasets during this process. For example, if we want to know if the dataset contains a Korean male aged 35 in New York with a rare disease A, we can only query twice. The first query is “Obtain the average height of all records.” The second query is “Obtain the average height of all records except 35-year-old + male + live in New York + rare disease A + Korea.” We can simply compare the results of different queries. If the results are inconsistent, we know that “Korean male aged 35 in New York with rare disease A” is in the dataset. DP is the algorithm to protect privacy from such multiple queries.
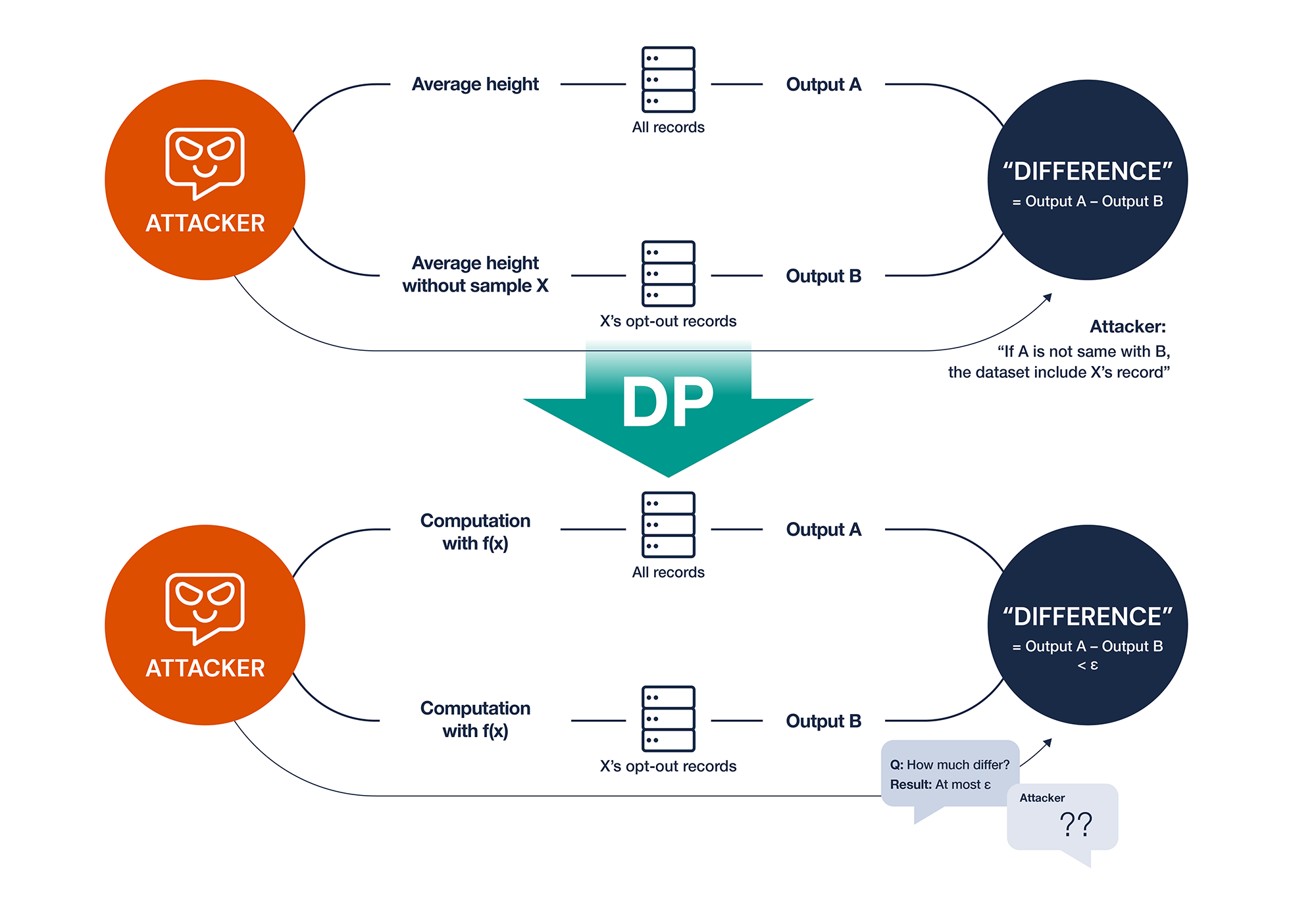
Figure 3. Illustration of scenarios before and after applying differential privacy
Figure 3 shows that X’s opt-out records are the records that exclude only X. From the standpoint of privacy attacker, attacker may query to exclude X records by including a condition that X may not have. If the queried value does not differ after adding the condition without X’s attributes, the returned records must not include X. This is a shortcoming of deanonymized data in public databases. Despite the fact that we remove a set of direct identifiers, privacy attacker can know if a dataset has one’s record by multiple queries which contain conditions one may not have.
This attack can be mitigated by DP. By altering the query output, the attacker is unable to determine whether the database contains one’s records. To achieve this, we need to change the statistics from each query. However, it is also necessary to determine how much noise adds to the statistics. The statistics would be rendered useless if more noise was introduced. On the other hand, adding minor noise raises the risk of invasion of privacy. As a result, determining optimal noise is critical. DP employs a specific distribution known as the Laplace distribution to determine noise size for not too large or small noise. Therefore, even if we ran the same query, we would get different results. There is research in genetics that suggests using test statistics from DP-applied queries in genome-wide association studies.
Conclusion
As the integration of multiple datasets for a wide range of studies becomes more common, it raises serious privacy concerns. As previously stated, a brief introduction to the privacy preserving techniques is provided, as are the concepts of terms related to privacy. In practice, most data processing organizations employ encryption to safeguard their internal networks. Encryption and deidentification, however, serve a different purpose. Encryption is a method for protecting data that has been accidentally leaked. However, deidentification is a method of preventing data from reidentification in public data.
The technique we described to protect privacy is applicable to shared data. It cannot completely ensure privacy preservation in shared genomic data that has already been deidentified in accordance with regulations. Genomic information, which is one type of biometric information, cannot be altered. As a result, institutions must carefully process genomic data in order to preserve data. Data anonymization techniques such as k-anonymity and DP can be used to reduce privacy risks in publicly available data.
References
- Kim, H. H., Kim, B., Joo, S., Shin, S. Y., Cha, H. S., & Park, Y. R. (2019). Why do data users say health care data are difficult to use? A cross-sectional survey study. Journal of Medical Internet Research, 21(8), e14126.
- Ohm, P. (2009). Broken promises of privacy: responding to the surprising failure of anonymization. UCLA Law Review, 57, 1701.
- Near, J. P. & Abuah, C. (2021). Programming differential privacy, 1, https://uvm-plaid.github.io/programming-dp
- Clayton, E. W., Evans, B. J., Hazel, J. W. & Rothstein, M. A. (2019). The law of genetic privacy: applications, implications, and limitations. Journal of Law and the Biosciences, 6(1), 1-36.
- ISO, ISO/TS 25237, Health informatics—Pseudomymization, 2008
- ISO standard (ISO 29100:2011)
- IHE(Integrating the healthcare Enterprise), IHE IT Infrastructure Handbook, de-identification, 2014
- ISO/IEC 20889
Get exclusive rare disease updates
from 3billion.








