In Feb 2019, 3billion launched genetic testing services for rare disease diagnoses based on whole exome sequencing (WES) technology. Since then, more than 40,000 patients have been sequenced at 3billion and approximately 45% of these patients have completed their diagnostic journey with us by receiving a clear molecular diagnosis.
However, there are still 55% of these patients who remain undiagnosed. We can think of three main reasons for the undiagnosed: 1) patients do not have an underlying genetic condition, 2) the disease-causing gene is not discovered yet, and 3) the disease-causing variant cannot be identified by the current platform/technology.
One approach we took to overcome the last issue was launching whole genome sequencing (WGS) last spring. However, since WES is still oftentimes more easily accessible and preferred by the physicians/patients, we also looked into the possibility of improving our current WES test by increasing the variant calling accuracy within a subset of the genomic regions that are prone to coverage drop-out.
Hereby, we are excited to announce that we have made significant progress towards achieving this goal and successfully increased the depth-of-coverage for a substantial part of these regions. The improvements we made can be categorized into four main groups and we will share the details below.
1. The Mitochondrial Genome is now part of the capture.
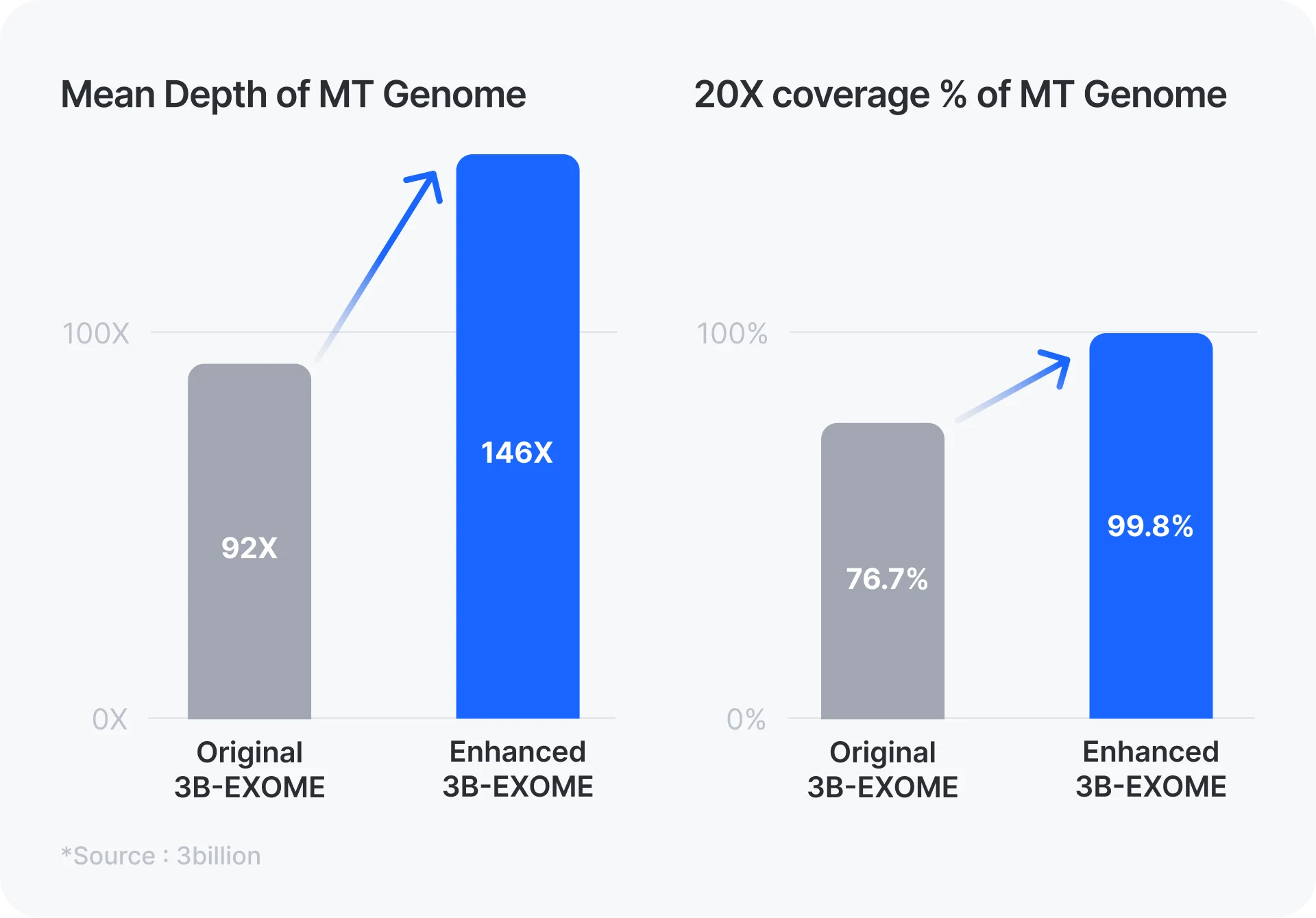
The exome capture kit we have been using did not include baits to capture the mitochondrial genome (MT) and therefore we had to rely on off-target sequencing reads from the MT. This resulted in a widely varying depth-of-coverage of the MT across samples and a large portion of the MT bases having very low (<20x) coverage within each sample. As you can see in the figure, with the enhanced whole exome sequencing (WES), the mean depth increased by ~70% and almost all MT bases consistently have >20x coverage now.
2. Improved Coverage of the RPGR ORF15 exon.
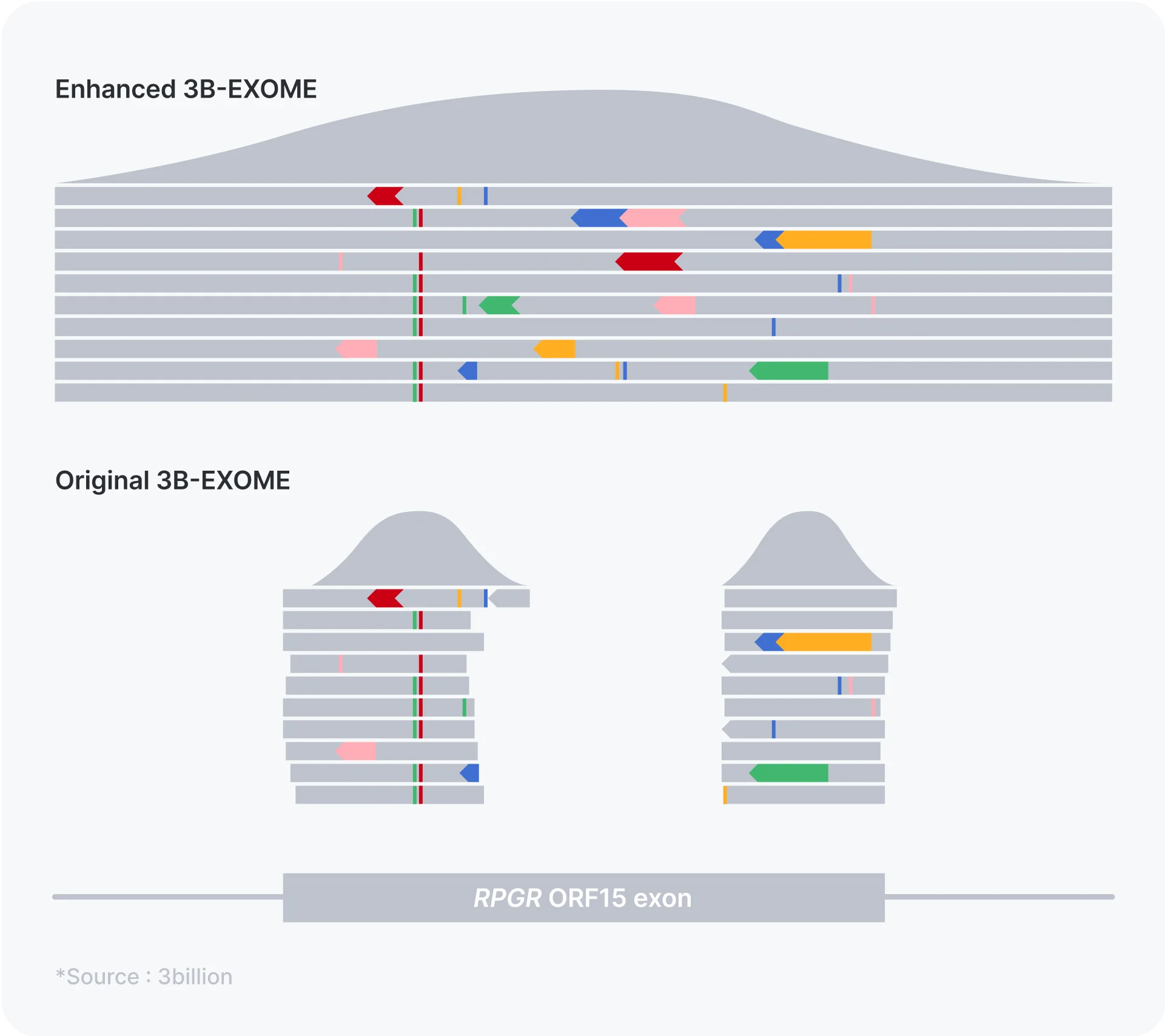
One of the exons of the RPGR gene, ORF15 exon, is a well-known exome coverage drop-out region despite many disease-causing variants being reported within. This imposed a risk of missing the diagnostic variant for patients presenting with retinitis pigmentosa, cone-rod dystrophy and/or macular degeneration. As you can see in the figure, with the enhanced 3B-EXOME, ORF15 exon is now completely sequenced with sufficient coverage.
3. Intronic regions of GLA and RPE65 genes are now covered.
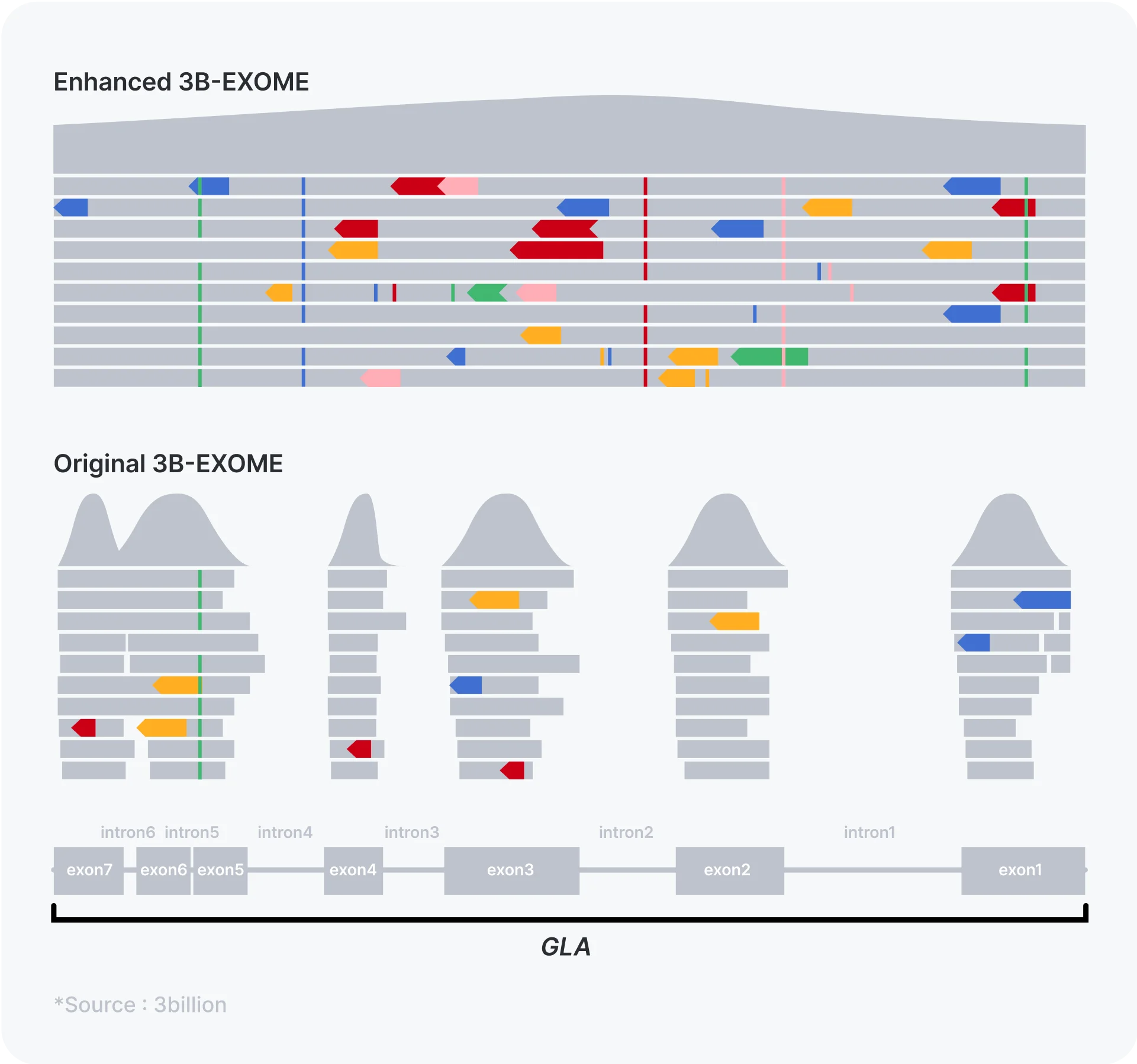
When it comes to rare diseases for which treatment options are available, an accurate molecular diagnosis is critical. Although most of the known disease-causing variants are reported within the coding exons, more and more intronic disease-causing variants resulting in splicing abnormalities are being identified as WGS and RNA sequencing are more frequently being performed. For the two genes, GLA and RPE65, that are associated with Fabry disease and Leber congenital amaurosis, respectively, we decided to capture all the introns as well to not only find the potentially disease-causing intronic variants but also breakpoints of small (<3 consecutive exons) copy number variants that would have been missed by the original 3B-EXOME.
4. Large number of non-coding disease-causing variant positions are covered.
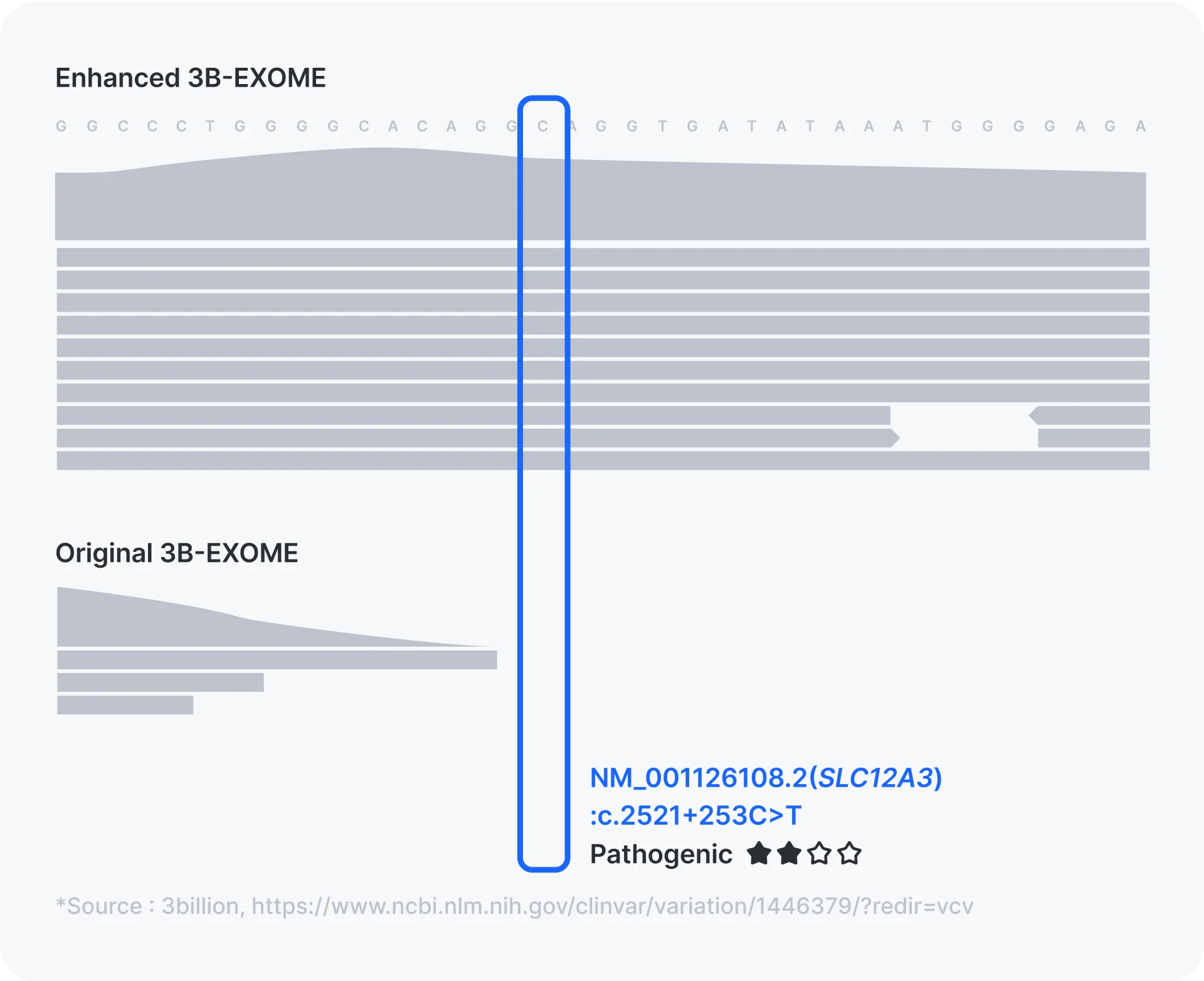
As mentioned above, with WGS and RNA sequencing becoming more accessible, more non-coding variants are being identified as disease-causing. Since WES only captures and sequences the exonic regions, deep intronic regions and untranslated regions reported to be disease-causing when mutated are not covered by default. We decided to target these variant positions by adding capture baits across these regions and we plan to regularly update the capture bait design for the newly identified non-coding disease causing variant regions. In this version of our Enhanced our WES, ~570 non-coding variant positions are captured and sequenced with sufficient coverage.
Hope you are convinced and as excited as we are that Enhanced 3B-EXOME has potential to find more disease-causing variants that would have been missed before. Enhanced 3B-EXOME will be applied to all samples arriving after May 1, 2023 and even better, the price will stay the same. We hope to share soon exciting cases that benefited from this enhancement. Please contact us for any related questions. We would be more than happy to discuss further details with you.