Those of you who have received 3billion genetic test results may have seen variants found in the patient described in HGVS nomenclature.
The concept of HGVS nomenclature is very familiar to those who work in the genomics field, but to those who do not, it may seem like an alien language. This is because it looks like a random mix of numbers and special characters. Though it may seem to be written in an indecipherable way, in fact, it accurately describes genetic variants according to strict rules and definitions.
Today I will discuss the rules for describing variants in HGVS nomenclature.
Which word to use, mutation or variant?
Before explaining HGVS nomenclature, let’s look at the background of the use of the word ‘variant’ to refer to any change that exists in the genome. Until the early 1990s, words such as ‘mutation’ and ‘polymorphism’ were used interchangeably. Mutation means (disease causing) changes and polymorphism means changes found with a frequency of 1% or more of the population, so words with different meanings were incorrectly used interchangeably. Furthermore, these two words had rather negative connotations, so it was recommended that the word ‘variant’ be used instead, because it is neutral and does not cause confusion in meaning, and this continues to this day.
What is HGVS nomenclature?
HGVS stands for Human Genome Variation Society and is the name of the most prominent international academic organization that studies the human genome. HGVS nomenclature refers to the genetic variant nomenclature recommended by the Human Genome Variation Society.
In 2000, HGVS first proposed rules and definitions for describing genetic variants, which have been gradually expanded and applied in various places, and today, they have become the global standard for describing genetic variants.
Understanding the rules used to write nomenclature
There are major rules to be followed in the HGVS nomenclature.
The rules and examples described in this section and the next section are partially extracted from the official site and paper, so please check them for more details.
- all variants should be described at the most basic level, the DNA level. Descriptions at the RNA and/or protein level may be given in addition.
- a letter prefix is mandatory to indicate the type of reference sequence used. Accepted prefixes are;
- “c.” for a coding DNA reference sequence
- “g.” for a linear genomic reference sequence
- “m.” for a mitochondrial DNA reference sequence
- “n.” for a non-coding DNA reference sequence
- “o.” for a circular genomic reference sequence
- “p.” for a protein reference sequence
- “r.” for an RNA reference sequence (transcript)
- descriptions at DNA, RNA and protein level are clearly different:
- DNA: 123456A>T : number(s) referring to the nucleotide(s) affected, nucleotides in CAPITALS using IUPAC-IUBMB assigned nucleotide symbols
- RNA: 76a>u: number(s) referring to the nucleotide(s) affected, nucleotides in lower case using IUPAC-IUBMB assigned nucleotide symbols
- Protein: Lys76Asn: the amino acid(s) affected in three- or one-letter code followed by a number IUPAC-IUBMB assigned amino acid symbols
- three-letter amino acid code is preferred (see Standards)
- the “*“ can be used to indicate the translation stop codon in both one- and three-letter amino acid code descriptions
- In HGVS nomenclature some characters have a specific meaning
- “+” (plus) is used in nucleotide numbering; c.123+45A>G
- “-” (minus) is used in nucleotide numbering; c.124-56C>T
- “” (asterisk) is used in nucleotide numbering and to indicate a translation termination (stop) codon (see Standards*); c.32G>A and p.Trp41
- “_” (underscore) is used to indicate a range; g.12345_12678del
- “[ ]” (square brackets) are used for alleles (see DNA, RNA, protein), which includes multiple inserted sequences at one position and insertions from a second reference sequence
- “;” (semi colon) is used to separate variants and alleles; g.[123456A>G;345678G>C] or g.[123456A>G];[345678G>C]
- “:” (colon) is used to separate the reference sequence file identifier (accession.version_number) from the actual description of a variant; NC_000011.9:g.12345611G>A
- “>” (greater than) is used to describe substitution variants (DNA and RNA level); g.12345A>T, r.123a>u (see DNA, RNA)
Specific abbreviations are used to describe different variant types.
- “>” is a substitution
- “del” is a deletion
- “dup” indicates a duplication; c.76dupA (see DNA, RNA, protein)
- “ins” indicates an insertion; c.76_77insG (see DNA, RNA, protein)
- duplicating insertions are described as duplications, not as insertions
- “inv” indicates an inversion; c.76_83inv (see DNA, RNA). Not used at protein level, usually described as “delins”
- “fs” indicates a frame shift; p.Arg456GlyfsTer17 (or p.Arg456Glyfs*17, see Frame shifts)
Put it into practice
Now that we know the dictionary meaning of each symbol and abbreviation, let's put the rules into practice.
- NC_000023.11:g.33344591del
The meaning of each part is as follows if we break it down into elements one by one.

This means that the sequence corresponding to the 33344591th in the genomic reference sequence of the reference database called NC_000023.11 has been deleted.
- Then shall we take a look at 3billion’s report?
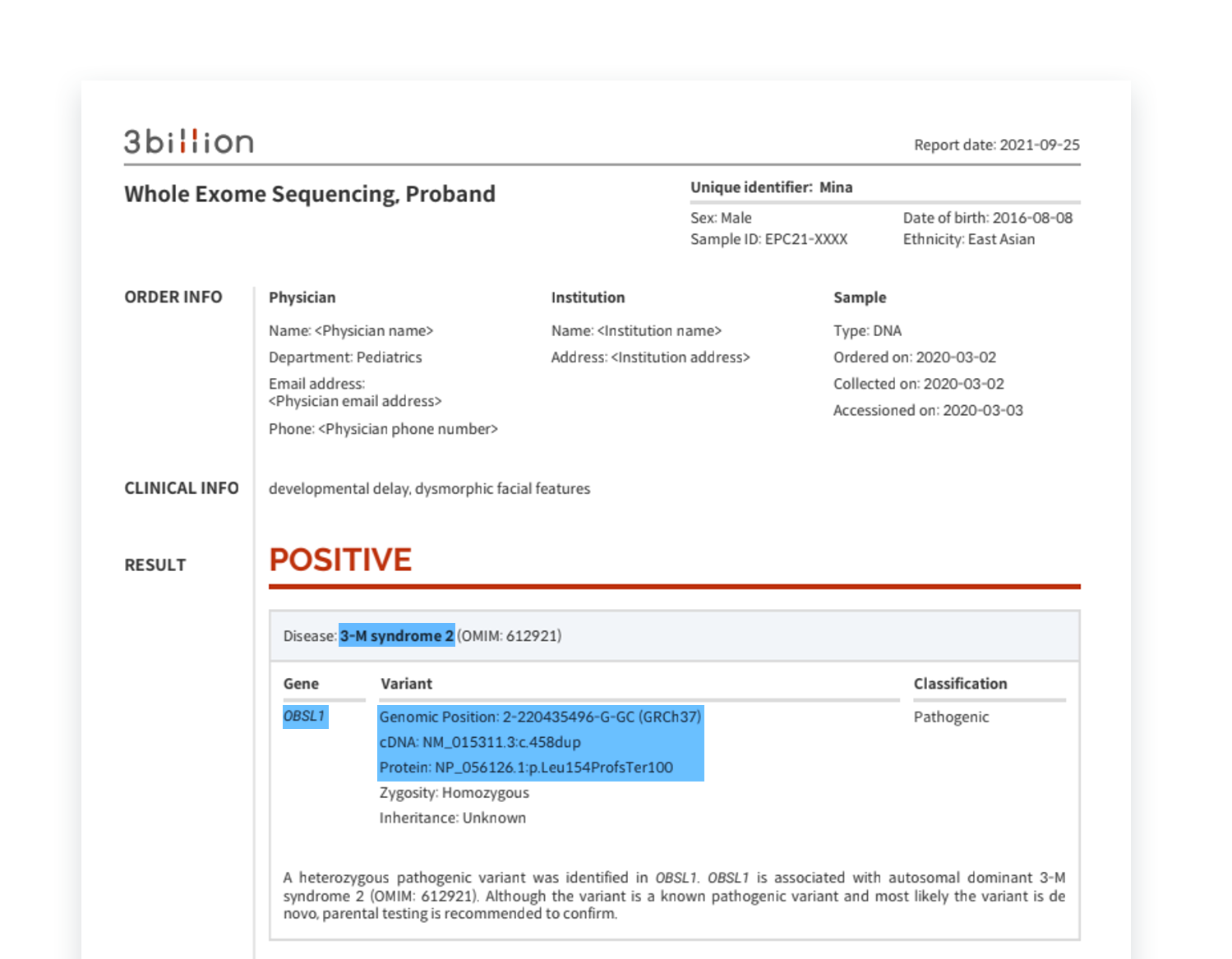
A pathogenic variant that appears to be the cause of 3-M syndrome 2 in the OBSL1 gene has been reported. Can you see that the variant is described in HGVS nomenclature in the Variant column? Let's interpret them one by one.
- Genomic Position: 2-220435496-G-GC (GRCh37)
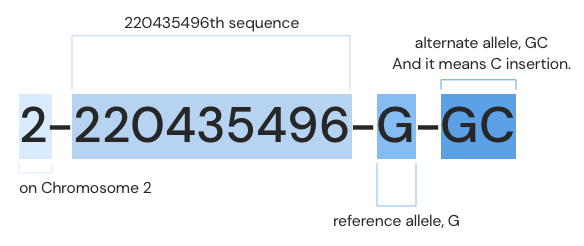
The genomic position is described according to the rules of “chromosome-position-reference allele-alternate allele”. Based on the GRCh37 reference sequence, it means that the 220435496th G sequence in the second chromosome has been changed to GC (that is, C is inserted).
- cDNA: NM_015311.3:c.458dup
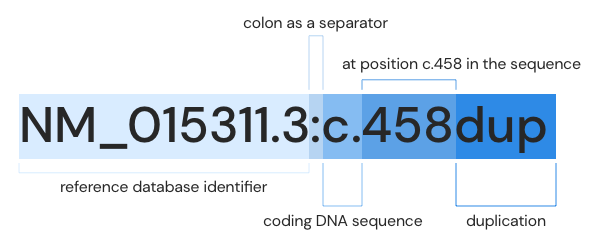
If we connect each block and translate it, it means that the 458th sequence has been duplicated from the coding DNA sequence of the reference database called NM_015311.3.
- Protein: NP_056126.1:p.Leu154Pro fs Ter100
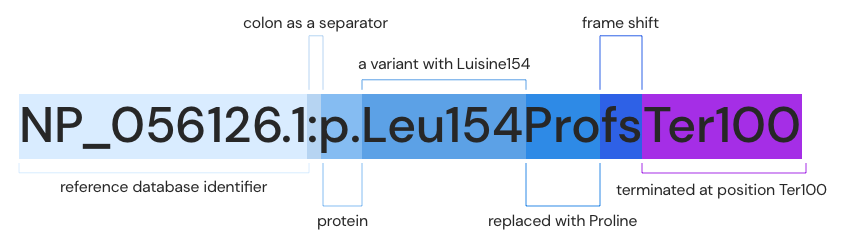
This describes changes at the protein level caused by changes in the cDNA sequence. Based on the NP_056126.1 reference database, Leucine154 was frameshifted to Proline, which means that protein translation was terminated at the Ter100 position.
In conclusion, this patient reported that the 220435496th G sequence on chromosome 2 was changed to GC, causing premature translational termination at the protein level, and this appears to be a pathogenic variant that causes a disease called 3-M syndrome 2.
Outro
It is not necessary to memorize all the abbreviations and special symbols presented by HGVS. If you become familiar with the format shown in examples above, I believe you will have no difficulty understanding HGVS nomenclature.
If you would like to read a genetic testing report with HGVS nomenclature, I think you will find the following interesting too: “New to genetic testing? Here are some practical tips on understanding whole exome sequencing test results.”
More detailed variant types and rules are presented in detail on the HGVS nomenclature official site, so if you want to learn more about them, I recommend that you visit the official site as well. I hope that this has helped you understand the concepts and rules of HGVS nomenclature.























